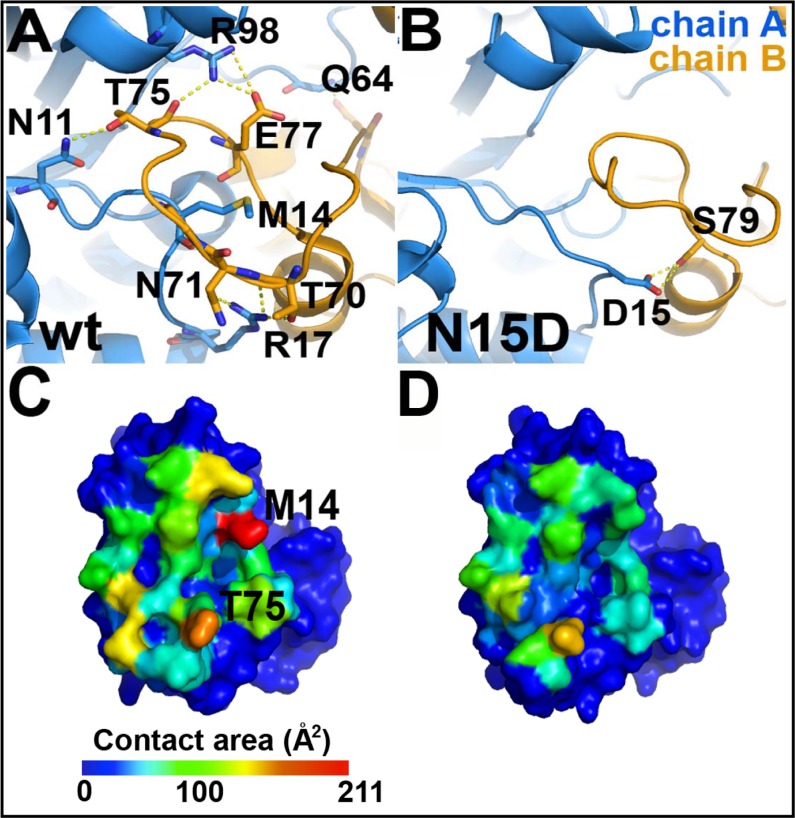
Fig 9. The dimeric interface is highly perturbed in the structure of the N15D mutant.
Panels A and C correspond to the WT HsTIM, whereas B and D correspond to the mutant N15D. (A) Hydrogen bonds in the interface of the wild-type enzyme that are lost in the N15D mutant (see S1 Table). The interactions were calculated with the HBplot server (http://dept.phy.bme.hu/virtuadrug/hbplot/bin/infopage.php). (B) There is a new intersubunit hydrogen bond in the N15D mutant that is not present in the wild type enzyme. This interaction is made between the mutated residue 15D and the side chain of S79. (C and D) Surface of a monomer interacting with the neighboring protein subunit. The total contact area per residue decreases from 2486 Å2 in the wild type enzyme (C) to 1561 Å2 on the N15D mutant (D) (see S2 Table), with the greatest decrease represented by residue M14 (red in C). These panels were drawn according to the results of the Contact Map Analysis server (http://ligin.weizmann.ac.il/cma/), which were plotted on the monomer surface according to the contact area of each residue on the interface on a blue (0 Å2) to red (211.8 Å2 for M14) scale basis.