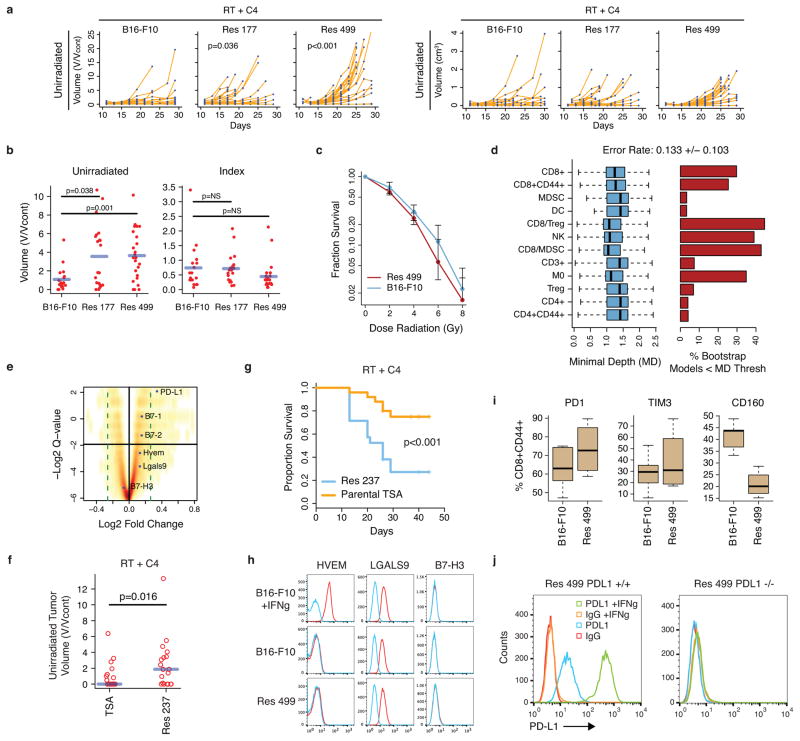
Extended Data Figure 2. Tumor cells resistant to RT + anti-CTLA4 upregulate PD-L1 but not other candidate inhibitory receptor pathways.
a) Unirradiated tumor growth (left: normalized, right: raw values) for mice implanted with Res 177 (n=21), Res 499 (n=25), and B16-F10 (n=18) melanoma cells and treated with RT + anti-CTLA4. For normalization, volumes were divided by average of untreated controls (V/Vcont) to account for differences in growth between untreated tumor types. The p-values are for comparisons with B16-F10 tumors. b) Corresponding tumor volumes of unirradiated or irradiated index tumors at day 21 (blue line is mean). c) Clonogenic survival for Res 499 and B16-F10 cells (n=2). d) Selection of immune variables that robustly predict resistance to RT + anti-CTLA4 using minimal depth (MD). A variable was selected if its MD was less than a threshold value for significance. Shown are bootstrap distributions of MD values (left) and % bootstrap models for which the MD for the indicated variable was significant (right). Bootstrap mean +/− SD for the out-of-bag prediction error rate is listed on top. e) Volcano plot of differentially expressed genes from resistant tumors. Horizontal black line is 5% false-discovery rate and dotted green line is fold-change cut-off. Ligands for select inhibitory receptors are indicated. See SI Table 1. f) Unirradiated tumor volumes (day 26–29) and g) survival after RT + anti-CTLA4 for mice with bilateral tumors from TSA breast cancer cells (n=25) or from the Res 237 subline selected to be resistant (n=21). h) Expression of candidate T cell inhibitory receptor ligands on B16-F10 and Res 499. Interferon-gamma (IFNg) responsiveness was tested. i) Boxplots show distribution of % positive CD8+CD44+ T cells for the indicated inhibitory receptor compared to IgG control. j) PD-L1 surface expression for CRISPR PD-L1 homozygous knockout Res 499 and wild type control cells. IFNg was used to induce PD-L1 and confirm abrogated response.