| TECHNIQUE | SCALE | ADVANTAGES | LIMITATIONS | REFERENCES |
|---|---|---|---|---|
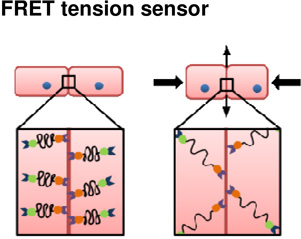 |
Molecular | + Ability to measure forces at molecular level in intact tissues | − Difficult validation process − Unable to obtain directional information − Molecular scale may not reflect cell- or tissue- scale tensions |
(Conway et al., 2013; Saeger et al., 2012) |
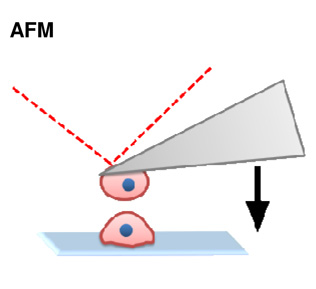 |
Molecular | + Ability to tether proteins and cells at nanometer resolution and under physiologica conditions. | − Receptor- ligand bond strengths may differ from in
vivo due to complex biological interactions. − Difficult to run experiments at high velocities due to viscous drag from cantilevers. |
(Puech et al., 2005) |
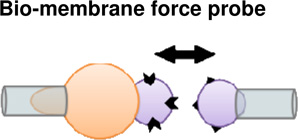 |
Molecular | + Detection of bond rupture forces with picoNewton sensitivity. + Micron sized trapped beads allow for more |
− Receptor- ligand bond strengths may differ from in
vivo due to complex biological interactions. − Limited rangel of detectable forces (large forces yield non-linear force- deformation Irelationships |
(Perret et al., 2004) (Bayas et al., 2006) |
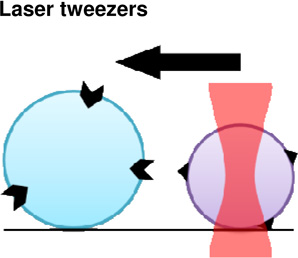 |
Molecular- Cell | + Isolation of single molecules + Precise control of force application. |
− Receptor- ligand bond strengths may differ from in
vivo due to complex biological interactions. − Requires optically homogeneous and highly purified samples. − Local heating of sample, potentially perturbing mechanics |
(Litvinov et al., 2002) |
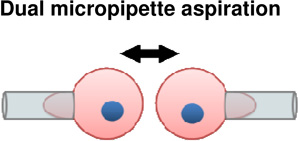 |
Cell | + Ability to grasp and move cells into contact with other cells or substrate | − Requires substantial deformation of cells. − Cells removed from native environment |
(Maitre et al., 2012) |
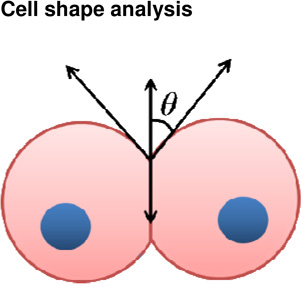 |
Cell | + Obtain cell surface tensions | − Dependent on contact time between cell pairs − Cells removed from native environment |
(Maitre et al., 2012) (Manning et al., 2010) |
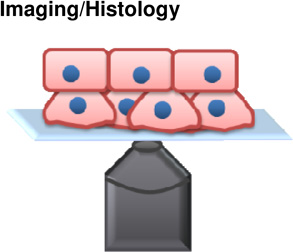 |
Cell-tissue | + Can be done in vivo + Genotype to phenotype relationship |
− Not mechanically quantitative − May change cell identity over time course of experiment |
(Kafer et al., 2007) (Dray et al., 2013) |
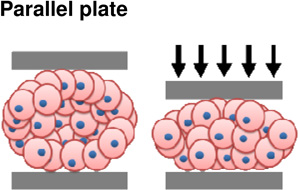 |
Tissue | + Directly measure tissue relaxation after application of known loads | − Requires spherical aggregates − Risk of changing cell identities over time course of experiment |
(Foty et al., 1994a) |
 |
Tissue | + Simple prep + Obtain whole tissue surface tension measureme nts |
− Requires spherical aggregates − Risk of changing cell identities over time course of experiment |
(Kalantarian et al., 2009) |

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.