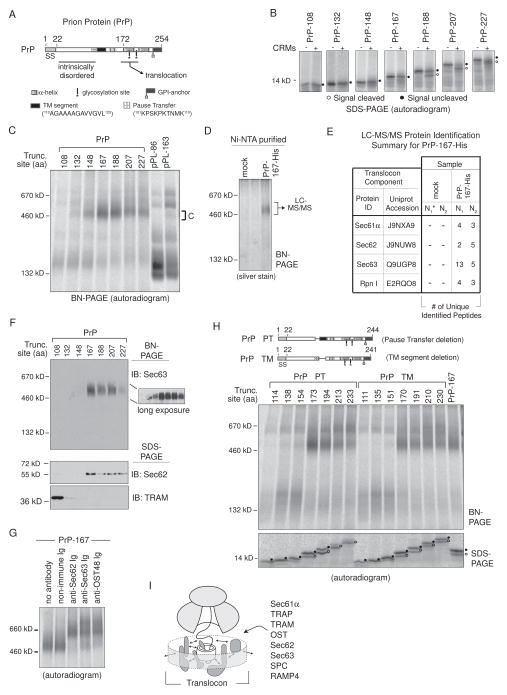
Figure 7. PrP Stabilizes Sec62 and Sec63 RAMP Complexes.
(A) A schematic of the prion protein (PrP) indicates location of signal sequence (SS), an N-terminal intrinsically disordered region, glycosylation sites, a GPI-anchor site, two helical domains, a pause-transfer sequence, and a TM segment. (B) SDS-PAGE analysis of translated PrP RTC intermediates ± CRMs. Peptidyl-tRNA was removed with RNase. (C) BN-PAGE of RAMPs prepared from indicated PrP and pPL translations. (D) Complex C isolation from PrP-167-His RAMP fractions by Ni-NTA purification after DTSSP crosslinking. (E) LC-MS/MS identified proteins from excised ~500 kD gel bands generated from mock or PrP-167-His Ni-NTA purified RAMP complexes as in (D). The asterisk denotes mock sample N1 that served as the control in a single experiment containing both pPL45-Zn-163-His (Figure 5E) and PrP-167-His samples. (F) Western blots of PrP RAMP fractions analyzed on BN- and SDS-PAGE. (G) Gel-shifts of PrP-167 RAMP complex C with indicated antibodies. (H) BN-PAGE of RAMPs prepared from indicated PrP truncations which lack the pause-transfer (PrP ΔPT) or TM segment (PrP ΔTM). (I) General model of dynamic translocon contacts (arrows) driven by nascent chain that could reflect repositioning of components within the RTC, recruitment of new factors, or affinity changes between bound components and/or polypeptide. See also Figure S5 and S7.