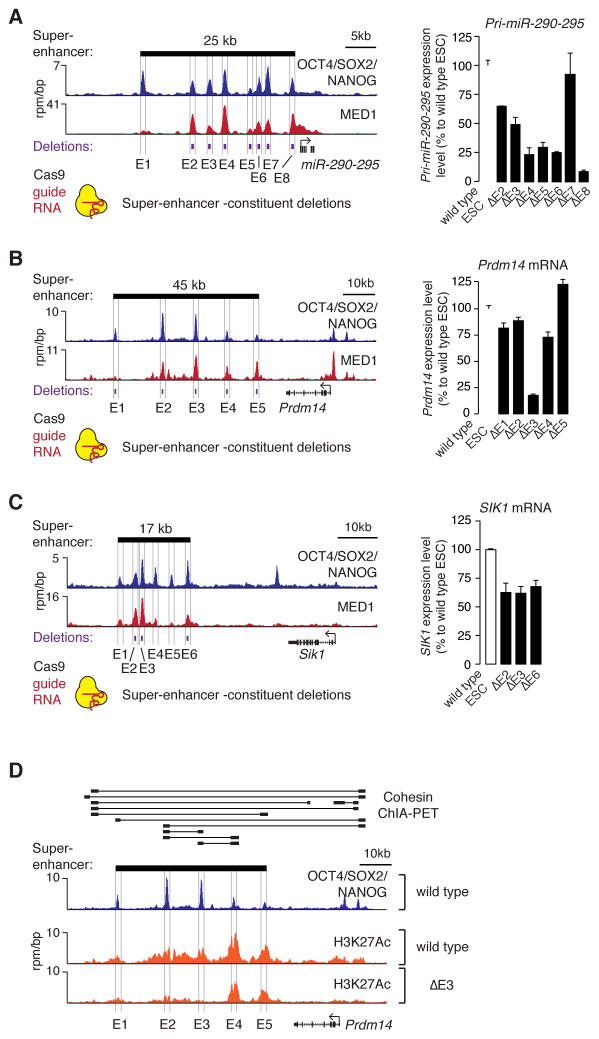
Figure 2. Contributions of super-enhancer constituents to gene expression in vivo.
(A) (left) ChIP-Seq binding profiles for OCT4, SOX2 and NANOG (merged) and Mediator (MED1) at the miR-290-295 locus in ESCs. (right) miR-290-295 expression level in ESCs in which the indicated super-enhancer constituents were deleted. Values correspond to mean +SD from three biological replicate experiments. (B) Gene expression analysis at the Prdm14 locus after deletion of super-enhancer constituents. (C) Gene expression analysis at the Sik1 locus after deletion of SE constituents. All values correspond to mean +SD from three biological replicate experiments. The difference of all values measured in deletion lines, except for miR-290-295 E7, are statistically significant compared to wild type (P<0.05, Student’s t-test). (D) ChIP-Seq binding profiles for H3K27Ac in wild type and Prdm14 E3 deleted ESCs. Cohesin (SMC1) ChIA-PET data for ESCs is shown above the binding profiles, where thick black bars connected by lines indicate regions that show high-confidence interactions (Dowen et al., 2014).
See also Table S2.