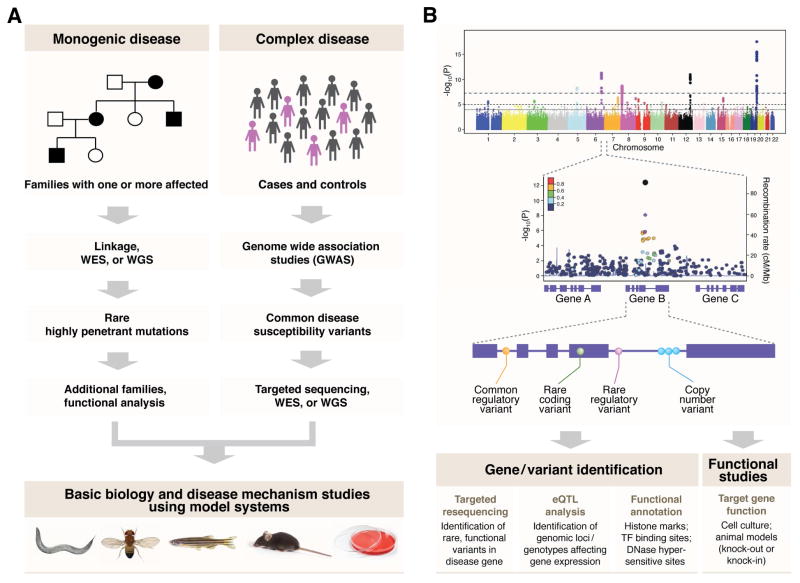
Figure 7.
General recommendations for genetic analysis of monogenic and complex diseases using next-generation approaches. A. Families with a monogenic (i.e., Mendelian) disease, even those with a small number of individuals, can be analyzed by whole exome or genome sequencing. In case of identification of mutation(s) in a novel gene, additional validation is required, including genetic analysis of more families/patients as well as functional analysis using animal models to understand the role in disease causation. For complex diseases, genome wide association study is performed on a large number of samples including patients and population-matched controls, to identify common susceptibility variants associated with the disease. Targeted sequencing, whole exome or genome sequencing can be applied to identify the causal variant at the susceptibility locus. Finally, multiple molecular genetic and cellular assays as well as studies in animal models are required to identify functional variants underlying trait association. WES, whole exome sequencing; WGS, whole genome sequencing, GWAS, genome wide association study. B. Schematic representation of follow-up on a GWAS hit to identify the causal gene/variant. Causal variants could be common regulatory variants, rare coding variants or copy number variants. Targeted sequencing around the associated locus can help identify the causal variant. Generally, more than one candidate gene is found at an associated locus, and eQTL analysis in disease relevant tissues can help identify the specific genotypes affecting gene expression within the causal gene. Additional functional annotation, including histone modifications, transcription factor binding sites and DNase I hypersensitivity sites can also help in identifying the causal gene. Ultimately, a high throughput functional assay in animal and/or cell-based models is highly recommended to understand the role of the gene in disease pathophysiology. cM/Mb, centimorgan (cM) per megabase pair (Mb); eQTL, expression quantitative trait locus, TF, transcription factor.