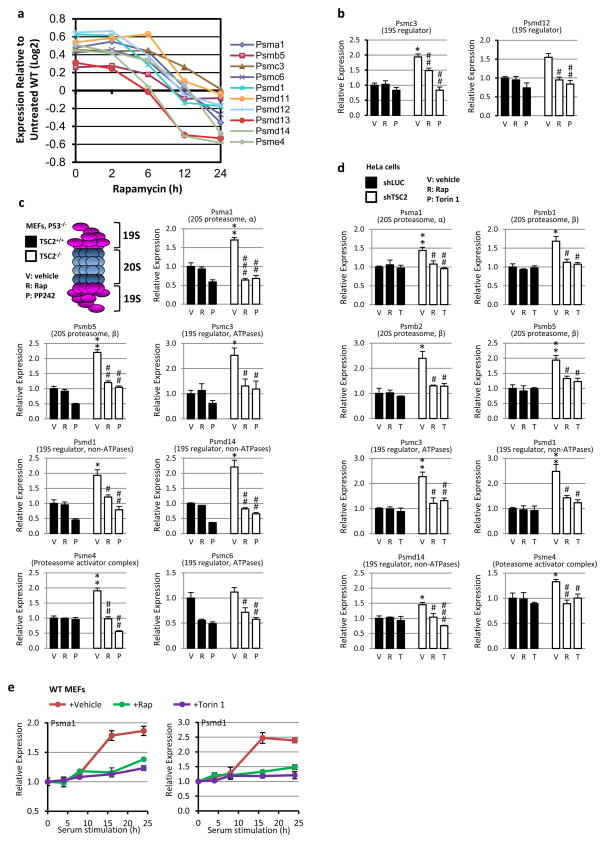
Extended Data Figure 3. mTORC1 signaling promotes PSM gene transcription.
a, PSM gene expression from a previous microarray experiment comparing expression in Tsc2−/− MEFs, over a time course of rapamycin treatment, to those in littermate Tsc2+/+ MEFs. Log2 expression levels provided are the average obtained from triplicate samples per time point of rapamycin treatment normalized to the expression levels in vehicle treated wild-type cells. b, The expression levels of two additional PSM genes from the experiment in Fig. 2a are shown. Data are mean ± s.e.m (n=3). *P<0.05 compared to vehicle-treated TSC2-expressing cells; #P<0.05, ##P<0.01 compared to vehicle-treated TSC2-deficient cells. c, The same experiment as Fig. 2a, except that PSM gene expression was analyzed in the same littermate-derived pair of Tsc2+/+ p53−/− and Tsc2−/− p53−/− MEFs use in a. Data are mean ± s.e.m. (n=3) relative to vehicle-treated TSC2+/+ cells. *p < 0.05 or **p < 0.01 compared to vehicle-treated TSC2+/+ cells; #p < 0.05, ##p < 0.01, or ###p < 0.001 compared to vehicle-treated TSC2−/− cells. d, The same experiment shown in Fig. 2a, except that PSM gene expression was analyzed in HeLa cells stably expressing shRNAs targeting firefly luciferase (shLUC) or those targeting human TSC2 (shTSC2). Data are mean ± s.e.m. (n=3) relative to vehicle-treated shLUC-expressing cells. *p < 0.05 or **p < 0.01 compared to vehicle-treated shLUC-expressing cells; #p < 0.05, ##p < 0.01, or ###p < 0.001 compared to vehicle-treated shTSC2-expressing cells. e, Cells were serum starved 16 h then stimulated with 10% serum in the presence of vehicle (DMSO), 20 nM rapamycin, or 250 nM Torin1. Transcript levels are shown as mean ± s.e.m. relative to vehicle (n=3). b–d, Statistical significance for pairwise comparisons evaluated with a two-tailed Student’s t test.