Abstract
Background. Persons with blood-stage Plasmodium falciparum parasitemia in the absence of symptoms are considered to be clinically immune. We hypothesized that asymptomatic subjects with P. falciparum parasitemia would differentially recognize a subset of P. falciparum proteins on a genomic scale.
Methods and Findings. Compared with symptomatic subjects, sera from clinically immune, asymptomatically infected individuals differentially recognized 51 P. falciparum proteins, including the established vaccine candidate PfMSP1. Novel, hitherto unstudied hypothetical proteins and other proteins not previously recognized as potential vaccine candidates were also differentially recognized. Genes encoding the proteins differentially recognized by the Peruvian clinically immune individuals exhibited a significant enrichment of nonsynonymous nucleotide variation, an observation consistent with these genes undergoing immune selection.
Conclusions. A limited set of P. falciparum protein antigens was associated with the development of naturally acquired clinical immunity in the low-transmission setting of the Peruvian Amazon. These results imply that, even in a low-transmission setting, an asexual blood-stage vaccine designed to reduce clinical malaria symptoms will likely need to contain large numbers of often-polymorphic proteins, a finding at odds with many current efforts in the design of vaccines against asexual blood-stage P. falciparum.
Keywords: malaria, systems biology, immunology, geographic medicine
Dogma in the malaria field holds that acquisition of clinical immunity to Plasmodium falciparum malaria results from the interaction of 2 factors. Specifically, the production of antigen-specific antibody [1, 2] and repeated parasite exposures over many transmission seasons are required for the onset of clinically protective immune responses as manifested by asymptomatic parasitemia [2], also called premonition [3]. These patterns have been characteristic of regions with high malaria transmission, primarily in sub-Saharan Africa [2, 4]. An exception to this was reported among previously malaria-naive adult transmigrants to Irian Jaya in the 1980s, who developed clinical immunity to P. falciparum within 2 years of arrival to this region of high malaria endemicity [5]. Previous studies using systems immunology/protein microarray experiments inferred antigen-specific immune responses by analyzing sera from residents of P. falciparum–endemic regions of Mali [4]. These data have been interpreted to indicate the concept that the development of acquired immunity is dependent on age and exposure to multiple infections, insofar as age can be considered a surrogate of clinical immunity and in areas where malaria infections are common and intense during annual rainy seasons [4].
Nonetheless, asymptomatic malarial parasite infections are common in Brazilian and Peruvian Amazonia, where transmission is considered to be hypoendemic and of low intensity [6–11]. Importantly, these well-established observations of asymptomatic parasitemia in Amazonia imply that clinical immunity may develop more rapidly in low-transmission settings [6–8, 12–15], which stands in contrast to high-transmission regions, where acquired immunity is manifested by asymptomatic parasitemia that takes years and intense seasonal or continuous transmission to develop [1, 4, 16, 17]. These observations imply that the immunological mechanisms underlying the acquisition of clinical immunity in areas with high as opposed to low transmission must be different and remain an unexplored yet high-priority area of the malaria field.
In the Amazon region, asymptomatic malaria parasitemia is common, and cases are not detected by passive case detection under normal surveillance, which conventionally relies on symptoms to drive diagnostic testing [8, 9]. Furthermore, clinically immune individuals are likely to be reservoirs of ongoing malaria transmission, and, perhaps just as important, understanding these acquired immune mechanisms may lead toward the development of new malaria vaccine strategies. In this context, there is evidence that antibody-dependent mechanisms play an important role in the reduction of parasitemia and can diminish clinical symptoms in humans, as was demonstrated by the passive transfer of hyperimmune immunoglobulin G (IgG) [1].
For this study, we used genome-scale protein microarrays to analyze the IgG responses to >800 recombinant proteins of asexual blood-stage P. falciparum in P. falciparum–infected subjects in the Peruvian Amazon. We hypothesized that asymptomatic subjects with P. falciparum parasitemia would differentially recognize a subset of P. falciparum proteins, compared with symptomatic patients, implying a contribution of these proteins to protective immune responses. Our objective was to compare antibody responses in symptomatic and asymptomatic individuals. We determined whether differentially recognized proteins in these 2 groups of malaria-affected subjects had undergone immune selection at a genomic level, to seek independent confirmation for a potential role in naturally acquired clinical immunity. Finally, we sought to identify the most immunodominant antigens shared by symptomatic and asymptomatic subjects, on a genomic scale, to identify novel candidate antigens for seroepidemiological surveillance.
METHODS
Ethics Statement
All patients provided written informed consent to be enrolled in this study, which was approved by the ethics committee of Universidad Peruana Cayetano Heredia (Lima, Peru) and by the directorate of health in the Loreto department of Peru. Written informed consent was obtained from each adult or, for children aged <18 years, from their parents or guardians.
Study Participants
A total of 38 plasma or serum samples from individuals infected with P. falciparum were collected during 2008–2012. The field activities of this study were performed in 17 different communities from the provinces of Maynas and Requena, Department of Loreto, Peru (Figure 1). Two groups of study subjects were recruited: (1) 24 individuals with symptomatic P. falciparum infection (10 females and 14 males; median age, 30 years; range, 9–59 years) and (2) 14 individuals with asymptomatic malaria (3 females and 11 males; median age, 31 years; range, 12–52 years).
Figure 1.
Map of the communities in the Loreto department of the northeastern Peruvian Amazon, where participants were enrolled. Most asymptomatic subjects were from the Alto Nanay district, which comprises the areas surrounding the Nanay, Pintuyacu, and Chambira rivers.
Symptomatic individuals were enrolled by passive case detection, all patients attended the San Juan Bautista Health Center in the District of San Juan received a diagnosis of P. falciparum infection on the basis of light microscopy findings. The microscopy-based diagnosis in all individuals enrolled was confirmed as a single-species (ie, P. falciparum) infection by real-time polymerase chain reaction (PCR) [18].
Most asymptomatic subjects were identified by active case detection after performing a diagnostic malaria survey in Atalaya and Diamante Azul, small villages located 6–7 km from Iquitos by motorboat in the district of Alto Nanay (Figure 1); this district had a malaria outbreak in March 2010.
The state of being asymptomatic (considered as a marker of indicate clinical immunity) was defined as the absence of malaria-referable symptoms during the 2 weeks before serum sampling for the study, including lack of fever and headache, pursuit of normal activities of daily living, and lack of any symptoms self-perceived to be indicative of malaria. Sera were obtained at the time when positive results were provided to patients, which was within 24 hours of performing microscopy of the blood smear. Any level of malaria parasitemia demonstrated by light microscopy in Peru mandates treatment, according to Peruvian Ministry of Health policy. Therefore, subjects with light microscopy–determined parasitemia were offered treatment when they received positive microscopy results.
Likewise, other asymptomatic individuals were enrolled following an index case-directed follow up strategy: once we identified a P. falciparum infection in a symptomatic individual during passive case detection, the index case's family or neighbors were visited weekly over the subsequent month to identify additional, asymptomatic cases, assuming a common risk factor of local malaria transmission. Some asymptomatic subjects had very low parasitemia that was detected only by real-time PCR, not by microscopy of a thick smear, of whom none developed fever or any malaria-related symptoms within 1 day of being identified as having parasitemia.
Individuals were excluded if they used antimalarial medication during the 2 weeks before enrollment and sample collection. No follow-up was done. Blood samples were collected before all individuals were treated for malaria with mefloquine/artesunate, following the National Drug Policy Guidelines of the Peruvian Ministry of Health.
Genome-Level P. falciparum Protein Microarray Analysis
A commercially available P. falciparum protein microarray was developed by Antigen Discovery (Irvine, California). Specifically, this study used a down-selected array based on the identification of seroreactive antigens from prior, larger array studies [19, 20], including an array of 2320 exon products (or “features”) corresponding to 1204 different genes [4, 21] and an array corresponding to 3126 different genes, or approximately 60% of the P. falciparum proteome (P. Felgner, unpublished data). The down-selected array in the present study is called “Pf824” and was designed to have 824 different P. falciparum features corresponding to 699 different P. falciparum genes [4]. The data reported here were obtained from probing the arrays and performing the confocal array scanning at the Universidad Peruana Cayetano Heredia, Lima, Peru; data analysis was done at the University of California–Irvine. To assess reproducibility, 4 asymptomatic and 12 symptomatic samples were reprobed at the University of California–Irvine, with comparable results. Signal detection was based on secondary antibody (biotin-conjugated goat anti-human IgG Fcy) and tertiary fluorescence detection (streptavidin-conjugated Alexa Fluor) accompanied by preblocking in 1% Escherichia coli lysate in blocking buffer. Slides were read on a GenePix 4200A confocal laser scanner, and spot intensity was analyzed using Pro ScanArray Express software.
The primary microarray data for this study have been deposited with Plasmodb.org and are available as Supplementary Table 1.
Data Analysis
Signal values from microarray scans were normalized by the vsn method and a Bayesian T test was used to identify differentially reactive antigens. Statistical analysis was performed in the R environment (http://www.r-project.org). Data were transformed by variance stabilization, using the vsn method from the Bioconductor suite (http://www.bioconductor.org) with spotted IgG-positive controls as normalization controls. Negative controls were based on empty vector transcription/translation reactions; these mean signal intensities were subtracted from protein detection signal intensities. A Bayesian-regularized t test adapted from Cyber-T for protein arrays [22] was used to compare the reactivity to the antigens between asymptomatic and symptomatic groups; the Benjamini–Hochberg method was used to correct for multiple measurements, estimate the false-discovery rate, and provide corrected P values. Proteins with transformed signal values of >2 standard deviations over the mean number of negative control signal values were considered reactive.
RESULTS
Characteristics of Study Participants
Symptomatic patients and asymptomatic carriers infected with P. falciparum were comparable by age, sex, and numbers of self-reported P. falciparum and Plasmodium vivax episodes (Table 1). All individuals reported at least 1 previous diagnosed P. falciparum infection (median, 2 infections [range, 1–4 infections] for symptomatic patients and 3 infections [range, 1–4 infections] for asymptomatic subjects; P = .011; Table 1). Levels of parasitemia were significantly higher (P < .001) in symptomatic patients (median, 7325 parasites/mL; range 71–39 778 parasites/mL) than in asymptomatic subjects (median, 526 parasites/mL; range, 24–12 915 parasites/mL), consistent with a previous report from the same area [9]. Half of participants reported working in agriculture; there were 6 minors (students), 5 housewives, 2 loggers, and 1 health promoter. The majority of studied individuals were men who stayed away from the village for >7 days because of work.
Table 1.
Demographic and Clinical Characteristics of Peruvian Amazon Subjects With a History of Plasmodium falciparum Infection
| Characteristic | Symptomatic (n = 24) | Asymptomatic (n = 14) | P Valuea |
|---|---|---|---|
| Age, y | 27 (9–59) | 32 (12–52) | .868 |
| Male sex | 58 | 79 | .205 |
| Episodes of self-reported malaria episodes, lifetime no. | |||
| With any Plasmodium parasite | 3 (1–8) | 4 (2–7) | .505 |
| With P. falciparum | 2 (1–4) | 3 (1–4) | .011 |
| With P. vivax | 2 (0–5) | 1.5 (0–4) | .325 |
| Parasitemia, parasites/µL | 7325 (71–39 778) | 526 (24–12 915) | <.001 |
Data are median value (range) or % of subjects.
a Continuous and binomial variables were compared between asymptomatic and symptomatic infection by the Mann–Whitney U test and χ2 test, respectively. Information for 5 individuals could not be obtained.
Differentially Reactive Antigens
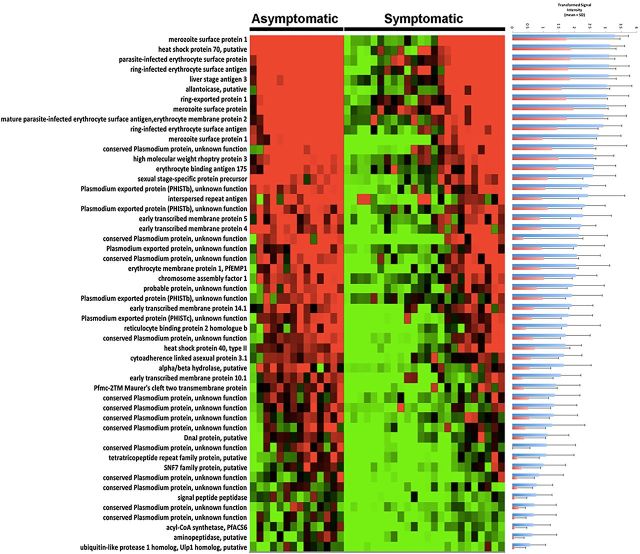
Sera from symptomatic and asymptomatic P. falciparum–infected subjects were used to probe P. falciparum protein microarrays, using previously described methods [4]. Fifty-two antigens from 51 proteins (PfMSP1 was divided into 2 segments) were differently reactive between samples from these 2 different patient groups (P ≤ .05, with the Benjamini–Hochberg correction for false-discovery rate; Figure 2 and Table 2). Of these antigens, 13 were identified as conserved Plasmodium proteins of unknown function. PfMSP1 (gene identification, PFI1475w), an established vaccine candidate, showed the highest reactivity among the differentially reactive antigens. Other vaccine candidates found were EBA175 (MAL7P1.176) and liver-stage antigen 3 (PFB0915w). Ring erythrocyte surface antigen (PFA0110) was also differentially reactive; this protein has been reported to be involved in modifying the biophysical properties of infected erythrocytes, and the effect seems to be enhanced by increasing the temperature to fever-associated levels [23, 24]. Two heat shock proteins, HSP40 (PFE0055ce4) and HSP70 (MAL7P1.228), were differentially recognized. Several other differentially recognized proteins are involved in interactions between infected red blood cells and both uninfected red blood cells and endothelial cells: REX1, ring exported protein 1 (PFI1735c) and early transcribed membrane proteins 4510.1 and 14.1 (PFD1120c, PFE1590w, PF10_0019, and PF14_0016).
Figure 2.
Plasmodium falciparum antigens found to be differently reactive between asymptomatic and symptomatic subjects. Red, highest signal intensity; green, lowest signal intensity. At right, variance-transformed values for asymptomatic subjects (blue bars) and symptomatic subjects (red bars), showing quantitatively different signal intensities. Gene identifications are specified in the same order as in Table 2. The heat map was made using MultiExperiment Viewer (MEV) 4.8.
Table 2.
Top 10 Differentially Reactive Plasmodium falciparum Antigens Recognized by Sera of Asymptomatic Versus Symptomatic Subjects With Malaria
| Name | Identifier |
|---|---|
| Merozoite surface protein 1 | PFI1475w |
| Heat shock protein 70, putative | Pf3D7_0831700 |
| Parasite:infected erythrocyte surface protein | PFE0060w |
| Ring:infected erythrocyte surface antigen | PFA0110w |
| Liver-stage antigen 3 | PFB0915w |
| Allantoicase, putative | PF14_0384e |
| Ring:exported protein 1 | PFI1735c |
| Merozoite surface protein 11 | PF10_0352 |
| Mature parasite:infected erythrocyte surface antigen, erythrocyte membrane protein 2 | PFE0040c |
| Ring:infected erythrocyte surface antigen | PF11_0509 |
Identification of Highly Reactive Antigens
A total of 535 proteins were significantly reactive with both groups of patient sera (Figure 3 and Table 3): 522 were identified in sera from asymptomatic subjects, and 471 were identified in sera from symptomatic patients. The sum of total IgG reactivity in relation to parasitemia was higher in asymptomatic parasitemic subjects, compared with symptomatic subjects. Although reactivity was higher in asymptomatic individuals, a set of 5 proteins was highly reactive for both clinical conditions, indicating the potential utility of these proteins in seroepidemiological studies. This set of broadly reactive antigens includes PF70 (PF10_0025) exo-antigen, unique to P. falciparum; 2 conserved proteins of unknown function (PF070053 and PFA0410), merozoite surface protein 10 (MSP10; PFF0995c), and a putative transcription factor (PF100075) with domains found in the apicomplexan AP2 family ([25].
Figure 3.
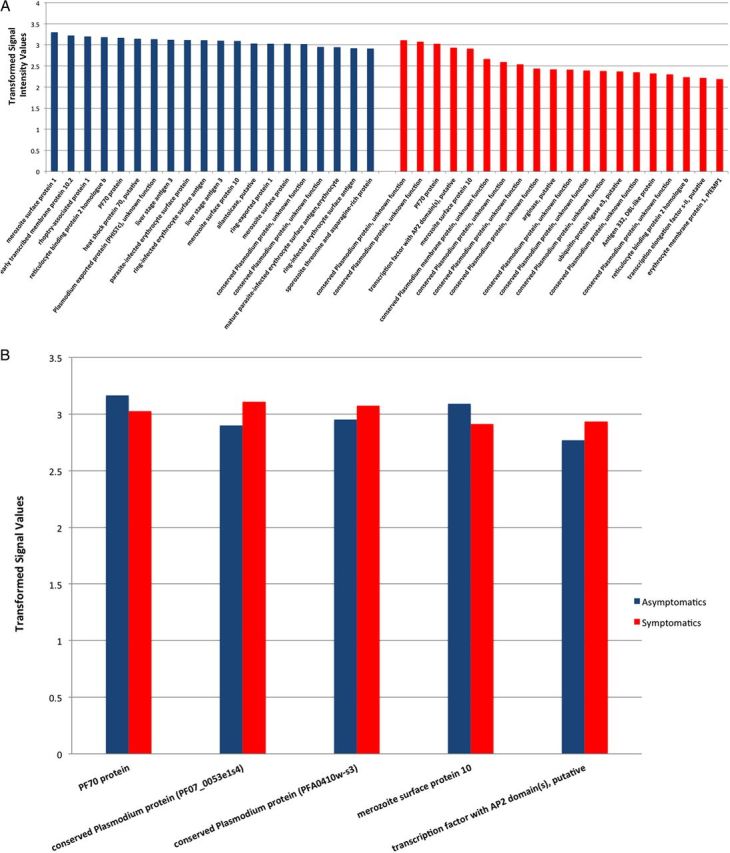
A, Plasmodium falciparum antigens most highly reactive by sera from asymptomatic (blue columns) and symptomatic (red columns) parasitemic subjects, ranked by signal intensity. These 2 sets of proteins have little overlap (see text). B, Seroepidemiological surveillance candidates, based on shared high reactivity by both asymptomatic and symptomatic subjects.
Table 3.
Top Reactive Antigens Recognized by Both Asymptomatic and Symptomatic Plasmodium falciparum-Infected Subjects Indicating Potential Use for Seroepidemiological Surveillance
| Name | Identifier | Asymptomatic | Symptomatic |
|---|---|---|---|
| Signal Intensity | Signal Intensity | ||
| PF70 protein | PF10_0025_2o2 | 59 663 | 44 981 |
| Conserved Plasmodium protein, unknown function | PF07_0053e1s4 | 52 571 | 47 710 |
| Conserved Plasmodium protein, unknown function | PFA0410w-s3 | 54 629 | 45 789 |
| Merozoite surface protein 10 | PFF0995c_1o1 | 57 564 | 44 053 |
| Transcription factor with AP2 domain(s), putative | PF10_0075e1s2 | 47 813 | 41 044 |
| Reticulocyte binding protein 2 homologue b | MAL13P1.176e1s2 | 60 450 | 32 694 |
| Merozoite surface protein 1 | PFI1475w-s1 | 64 353 | 29 430 |
| Early transcribed membrane protein 10.2 | PF10_0323e1s1 | 61 707 | 30 961 |
| Rhoptry-associated protein 1 | PF14_0102e1s1 | 60 665 | 31 447 |
| Liver stage antigen 3 | PFB0915w-e2s2 | 58 657 | 31 290 |
Enrichment of Genetic Diversity of Genes Encoding Differentially Reactive Antigens
The nonsynonymous genetic diversity (π) of genes encoding the set of differentially recognized P. falciparum antigens was compared with the coding sequences found in the P. falciparum genome as a whole (Table 4). The genes encoding the set of differentially recognized antigens had a significantly higher level of nonsynonymous single-nucleotide polymorphism diversity than the rest of the coding genes (mean, 2.6 × 10–3 vs 7.9 × 10–4; Mann–Whitney U test, W = 117 271, P = 8.05 × 10–5) in a collection of previously sequenced parasites from Senegal [26]. An analogous comparison of synonymous nucleotide polymorphisms found no significant enrichment of diversity in the differentially recognized gene set (W = 81 099, P = .29). These observations suggest positive immune selection for amino acid polymorphism in these antigens.
Table 4.
piN/piS* Ratios of Genes Encoding Differentially Recognized Plasmodium falciparum Proteins
| Locus | Identifier | piN | piS | piN/piS |
|---|---|---|---|---|
| PF3D7_0102200 | PFA0110w | 0.001515608 | 0.002313864 | 0.655011615 |
| PF3D7_0202400 | PFB0115w | 0.00069447 | 0.000656881 | 1.057223467 |
| PF3D7_0220000 | PFB0915w | 0.003143646 | 0.004420504 | 0.711150975 |
| PF3D7_0301900 | PFC0095c | 0.000676163 | 0.002197125 | 0.307748959 |
| PF3D7_0302500 | PFC0120w | 0.007704959 | 0.027030855 | 0.285043101 |
| PF3D7_0316500 | PFC0720w | 4.00E-05 | 0 | NDa |
| PF3D7_0401900 | PFD0085c | 0.002711623 | 0.001314006 | 2.063630592 |
| PF3D7_0406200 | PFD0310w | 0.000126178 | 0 | NDa |
| PF3D7_0423700 | PFD1120c | 0.00242942 | 0 | NDa |
| PF3D7_0424800 | PFD1180w | 0.0004852 | 0.000379997 | 1.276850879 |
a Not determined (ND), because the gene has no synonymous single-nucleotide variants.
* piN/piS indicates ratio of nonsynonymous to synonymous single nucleotide polymorphisms.
DISCUSSION
Using a genome-scale protein microarray to analyze naturally acquired human antibody responses to P. falciparum asexual blood-stage proteins, we demonstrate that a simple case definition of asymptomatic P. falciparum parasitemia robustly predicted immune responses associated with naturally acquired clinical immunity to malaria. A key aspect of this study is that we carefully defined asymptomatic parasitemia (and hence clinical immunity) on an individual basis in our study population, rather than using age as a proxy for clinical immunity in very large study populations as previous studies using P. falciparum protein microarray analysis have done [4]. A potential caveat to consider from the data presented here is that parasite density as a predictor of lack of symptoms could be considered as a potential confounder. It has long been known that low parasite density is associated with lack of malaria-related fever in the malaria-holoendemic setting. The data presented here support the hypothesis that a specific antibody profile produced by the protein microarray analysis predicts lack of symptoms and hence a state of clinical immunity. It would be ideal to compare genomic-level antibody responses in symptomatic and asymptomatic patients with higher parasite densities than found in the patients described here. However, in the Peruvian Amazon region, asymptomatic P. falciparum–infected individuals with high parasite densities are not found, so such an analysis is not currently feasible.
In this study, we found that asymptomatically infected subjects had antibody responses that differentially recognized 51 P. falciparum protein antigens from a panel of 824 proteins previously determined to be a general set of all P. falciparum asexual stage proteins recognized globally by human IgG immune responses [4, 19, 27]. The conclusions provided the protein microarray analysis are strongly supported by the finding that genes encoding differentially reactive antigens demonstrated evidence of immune selection on a genomic scale, as evidenced by statistically significant enrichment of the genes encoding differentially reactive proteins as harboring nonsynonymous variations, compared with the genome as a whole.
Previously identified immunogenic antigens, including vaccine candidates such as MSP1 [2], have been found to have the highest reactivity, as previously found using other approaches [28–31]. In addition, new hitherto unstudied proteins not previously recognized as potential vaccine candidates demonstrated high differential reactivity (Figure 2 and Table 2), suggesting that protective immunity against malaria parasites regardless of stage will be complex and likely dependent on simultaneous immune responses against large sets of P. falciparum antigens, as previously described for antisporozoite protective immunity [21].
In recent years, the recognition that asymptomatic Plasmodium infection is a critical barrier to malaria elimination has begun to guide new strategies in control strategies. The present analysis identifies new potential P. falciparum antigens that may be useful for large-scale seroepidemiological surveys of regions and microgeographic areas of malaria transmission. Subclinical parasitemia plays a key role in the maintenance of continuing malaria transmission and represents an important potential for regional reintroduction of malaria [32–34]. Hence, the development of tools to enable rapid, efficient, and accurate detection and estimation of asymptomatic parasitemia in malaria-endemic populations and at key portals of entry (in relation to interisland or intraregional human mobility) is a priority for new malaria policy makers. The study design described here, combined with data obtained using a genomic-level analytical tool such as the P. falciparum asexual blood-stage-based protein microarray, provides a road map to the development of such seroepidemiological tools. Work is ongoing to determine the kinetics and age-stratification of IgG responses to broadly recognized P. falciparum antigens after drug treatment.
This study, in the low-transmission region of the Peruvian Amazon, is the first to deploy this new high-throughput screening system. We were able to identify proteins as potential targets of protective immunity and to identify such proteins as potential markers for seroepidemiological studies to identify malaria transmission patterns on a population basis. The data suggest that naturally occurring clinical immunity—and, by extension, an efficacious malaria vaccine targeting blood-stage parasites—should target larger groups of key antigens differentially recognized by asymptomatic individuals, compared with symptomatic individuals. By taking advantage of having identified the most immunodominant proteins recognized by both symptomatic and asymptomatic patients on a genomic scale, this approach identifies new markers that can be used to differentiate asymptomatic and symptomatic subjects. It emerges from these and similar past results that protection to infection and symptoms comes from reaction to a broad set of proteins, a fact that may go against current vaccination strategies focused on single protein subunit vaccines.
Because of some limitations, this study needs to be replicated in different malaria-endemic regions and with larger sample sizes. A bigger population sample would likely increase the variety of antigens to which response is involved in developing clinical immunity. Nonetheless, rigorous statistical analysis identified a subset of 51 differentially reactive protein antigens associated with the state of premonition; while a larger subject population would likely identify more differentially reactive antigens, the groups under study nonetheless yielded robust information. Further, the independent finding that the genes encoding the differentially reactive antigens were significantly enriched for nonsynonymous mutations, compared with the genome as a whole, strongly supports our conclusion that the differentially reactive antigens collectively indicate protective immune responses and, hence, immune selection for amino acid polymorphisms in this set of proteins. Another observation that remains to be confirmed is the finding that the set of protein antigens recognized by the asymptomatic subjects appears to have 2 patterns of seroreactivity (Figure 2), despite statistical analysis demonstrating that all subjects in this group recognized the same group of 51 differentially reactive protein antigens. It seems unlikely that these infections were simply early infections about to become symptomatic, given the case enrollment criteria and previous experience with similar populations in this region [6, 8, 12]. There were no apparent phenotypic differences in responses to infection within the asymptomatic patient group. Finally, one last potential concern is that the protein microarray comprised recombinant P. falciparum proteins produced using a cell-free E. coli–based protein expression system that may not produce conformationally correct, properly folded proteins. The antibody reactivity detected in the arrays reported here resulted in a robust correlation between antibody reactivity to proteins expressed in this system and to the corresponding well-characterized correctly folded recombinant proteins spotted on the same array, as previously described [4, 21, 27, 35]. Further work is necessary to confirm the discovery approach used here, using properly folded, recombinant top-hit proteins in an enzyme-linked immunosorbent assay format. Such analysis will likely be even more revealing when large-scale protein arrays produced with eukaryotic-system-produced recombinant proteins are used.
PfMSP10, a previously unstudied protein, was identified as highly reactive in symptomatic and asymptomatic parasitemic subjects. This protein, which has been suggested to be undergoing purifying selection in P. vivax [36], has predicted structural characteristics similar to that of other merozoite surface proteins (eg, MSP1 and MSP8) in that it is predicted to have 2 tandemly arrayed epidermal growth factor (EGF)–like domains at the C terminus. These domains, as with other EGF domain–containing proteins [37–40], require conformationally dependent antibodies as targets of protective immunity [41]. Nonetheless, no previous study has suggested a role in clinical immunity of PfMSP10 in P. falciparum, nor do the data here do so, because this protein, while highly reactive with antibodies from naturally infected humans, is not differentially recognized between symptomatic and clinically immune individuals. Therefore, data presented here suggest that PfMSP10 might serve a useful function in seroepidemiology studies to identify populations in which malaria transmission is occurring, because the protein microarray analysis indicates that this protein is strongly recognized by antibodies found in both asymptomatic and symptomatic individuals. Similar results have been found when analyzing samples from individuals infected with P. vivax (R. Chuquiyauri et al, unpublished data).
In the Peruvian Amazon, studies using classic approaches have shown a robust IgG response against primarily MSP1–19 and were sufficient to produce a positive anti-MSP1–19 IgG response for >5 months [15, 42]. These data were interpreted as indicating protective immunity. In contrast, a recent report provided strong evidence that the exposed population present in a low-transmission setting has a capacity to develop clinical immunity (as manifested by low-level parasitemia in the absence of malaria-referable symptoms) that is likely associated, at least in part, with antibody responses to defined P. falciparum invasion ligands [43].
In contrast to single-antigen-based serological analysis based on vaccine candidates such as MSP1, genome-based proteome microarray analysis provides highly granular information with important potential for unbiased identification of antigenic correlates of protective immunity. To infer such conclusions requires robust technology combined with careful clinical and parasitological phenotyping. The data reported here implicate a large but limited set of antibody responses to asexual blood-stage P. falciparum antigens as contributing key immune protection in malaria, taking advantage of naturally acquired infection in a low-transmission region of the Peruvian Amazon. These observations need to be replicated in other comparable settings for external validation, but the approach described here is robust, with a high potential to lead to novel approaches to malaria vaccine design, as well as seroepidemiological surveillance.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank Paula Maguina for expert and essential scientific, ethical, and logistical contributions that allowed this work to come to fruition.
Financial support. This work was supported by the National Institutes of Health (grants U19AI089681, 1R01AI067727, K24AI068903, and D43TW007120 to J. M. V. and grants R01AI05759206, AI075692, and R01RHLO86488 to P. F.).
Potential conflicts of interest. P. F. has an equity interest in Antigen Discovery, which is developing products related to the research described in this article; and serves on the advisory board of Antigen Discovery and receives compensation for these services. The terms of this arrangement have been reviewed and approved by the University of California in accordance with its conflict of interest policies. D. M. M. is an employee of Antigen Discovery. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Cohen S, McGregor IA, Carrington S. Gamma-globulin and acquired immunity to human malaria. Nature 1961; 192:733–7. [DOI] [PubMed] [Google Scholar]
- 2.Crompton PD, Moebius J, Portugal S, et al. Malaria immunity in man and mosquito: insights into unsolved mysteries of a deadly infectious disease. Ann Rev Immunol 2014; 32:157–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sergent E, Parrot L. Premunition in bovine piroplasmosis and human malaria. Ann Trop Med Parasitol 1950; 44:329–30. [DOI] [PubMed] [Google Scholar]
- 4.Crompton PD, Kayala MA, Traore B, et al. A prospective analysis of the Ab response to Plasmodium falciparum before and after a malaria season by protein microarray. Proc Natl Acad Sci U S A 2010; 107:6958–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baird JK, Jones TR, Danudirgo EW, et al. Age-dependent acquired protection against Plasmodium falciparum in people having two years exposure to hyperendemic malaria. Am J Trop Med Hyg 1991; 45:65–76. [DOI] [PubMed] [Google Scholar]
- 6.Alves FP, Durlacher RR, Menezes MJ, Krieger H, Silva LH, Camargo EP. High prevalence of asymptomatic Plasmodium vivax and Plasmodium falciparum infections in native Amazonian populations. Am J Trop Med Hyg 2002; 66:641–8. [DOI] [PubMed] [Google Scholar]
- 7.Vinetz JM, Gilman RH. Asymptomatic Plasmodium parasitemia and the ecology of malaria transmission. Am J Trop Med Hyg 2002; 66:639–40. [DOI] [PubMed] [Google Scholar]
- 8.Roshanravan B, Kari E, Gilman RH, et al. Endemic malaria in the Peruvian Amazon region of Iquitos. Am J Trop Med Hyg 2003; 69:45–52. [PubMed] [Google Scholar]
- 9.Branch O, Casapia WM, Gamboa DV, et al. Clustered local transmission and asymptomatic Plasmodium falciparum and Plasmodium vivax malaria infections in a recently emerged, hypoendemic Peruvian Amazon community. Malar J 2005; 4:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chuquiyauri R, Paredes M, Penataro P, et al. Socio-demographics and the development of malaria elimination strategies in the low transmission setting. Acta Trop 2012; 121:292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.da Silva-Nunes M, Moreno M, Conn JE, et al. Amazonian malaria: Asymptomatic human reservoirs, diagnostic challenges, environmentally driven changes in mosquito vector populations, and the mandate for sustainable control strategies. Acta Trop 2012; 121:281–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camargo EP, Alves F, Pereira da Silva LH. Symptomless Plasmodium vivax infections in native Amazonians. Lancet 1999; 353:1415–6. [DOI] [PubMed] [Google Scholar]
- 13.Alves FP, Gil LH, Marrelli MT, Ribolla PE, Camargo EP, Da Silva LH. Asymptomatic carriers of Plasmodium spp. as infection source for malaria vector mosquitoes in the Brazilian Amazon. J Med Entomol 2005; 42:777–9. [DOI] [PubMed] [Google Scholar]
- 14.Baird JK, Fryauff DJ, Hoffman SL. Primaquine for prevention of malaria in travelers. Clin Infect Dis 2003; 37:1659–67. [DOI] [PubMed] [Google Scholar]
- 15.Clark EH, Silva CJ, Weiss GE, et al. Plasmodium falciparum malaria in the Peruvian Amazon, a region of low transmission, is associated with immunologic memory. Infect Immun 2012; 80:1583–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGregor IA. Mechanisms of acquired immunity and epidemiological patterns of antibody responses in malaria in man. Bull WHO 1974; 50:259–66. [PMC free article] [PubMed] [Google Scholar]
- 17.Langhorne J, Ndungu FM, Sponaas AM, Marsh K. Immunity to malaria: More questions than answers. Nat Immunol 2008; 9:725–32. [DOI] [PubMed] [Google Scholar]
- 18.Mangold KA, Manson RU, Koay ES, et al. Real-time PCR for detection and identification of Plasmodium spp. J Clin Microbiol 2005; 43:2435–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doolan DL, Aguiar JC, Weiss WR, et al. Utilization of genomic sequence information to develop malaria vaccines. J Exp Biol 2003; 206:3789–802. [DOI] [PubMed] [Google Scholar]
- 20.Riehle MA, Srinivasan P, Moreira CK, Jacobs-Lorena M. Towards genetic manipulation of wild mosquito populations to combat malaria: advances and challenges. J Exp Biol 2003; 206:3809–16. [DOI] [PubMed] [Google Scholar]
- 21.Trieu A, Kayala MA, Burk C, et al. Sterile protective immunity to malaria is associated with a panel of novel P. falciparum antigens. Mol Cell Proteomics 2011; 10:M111.007948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 2001; 17:509–19. [DOI] [PubMed] [Google Scholar]
- 23.Diez-Silva M, Park Y, Huang S, et al. Pf155/RESA protein influences the dynamic microcirculatory behavior of ring-stage Plasmodium falciparum infected red blood cells. Sci Rep; 2:614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silva MD, Cooke BM, Guillotte M, et al. A role for the Plasmodium falciparum RESA protein in resistance against heat shock demonstrated using gene disruption. Mol Microbiol 2005; 56:990–1003. [DOI] [PubMed] [Google Scholar]
- 25.Sinha A, Hughes KR, Modrzynska KK, et al. A cascade of DNA-binding proteins for sexual commitment and development in Plasmodium. Nature 2014; 507:253–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park DJ, Lukens AK, Neafsey DE, et al. Sequence-based association and selection scans identify drug resistance loci in the Plasmodium falciparum malaria parasite. Proc Natl Acad Sci U S A 2012; 109:13052–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doolan DL, Mu Y, Unal B, et al. Profiling humoral immune responses to P. falciparum infection with protein microarrays. Proteomics 2008; 8:4680–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rotman HL, Daly TM, Long CA. Plasmodium: immunization with carboxyl-terminal regions of MSP-1 protects against homologous but not heterologous blood-stage parasite challenge. Exp Parasitol 1999; 91:78–85. [DOI] [PubMed] [Google Scholar]
- 29.Keitel WA, Kester KE, Atmar RL, et al. Phase I trial of two recombinant vaccines containing the 19kd carboxy terminal fragment of Plasmodium falciparum merozoite surface protein 1 (msp-1(19)) and T helper epitopes of tetanus toxoid. Vaccine 1999; 18:531–9. [DOI] [PubMed] [Google Scholar]
- 30.Chenet SM, Branch OH, Escalante AA, Lucas CM, Bacon DJ. Genetic diversity of vaccine candidate antigens in Plasmodium falciparum isolates from the Amazon basin of Peru. Malar J 2008; 7:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barry AE, Trieu A, Fowkes FJ, et al. The stability and complexity of antibody responses to the major surface antigen of Plasmodium falciparum are associated with age in a malaria endemic area. Mol Cell Proteomics 2011; 10:M111.008326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beshir KB, Sutherland CJ, Sawa P, et al. Residual Plasmodium falciparum parasitemia in Kenyan children after artemisinin-combination therapy is associated with increased transmission to mosquitoes and parasite recurrence. J Infect Dis 2013; 208:2017–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bousema T, Drakeley C. Epidemiology and infectivity of Plasmodium falciparum and Plasmodium vivax gametocytes in relation to malaria control and elimination. Clin Microbiol Rev 2011; 24:377–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sawa P, Shekalaghe SA, Drakeley CJ, et al. Malaria transmission after artemether-lumefantrine and dihydroartemisinin-piperaquine: a randomized trial. J Infect Dis 2013; 207:1637–45. [DOI] [PubMed] [Google Scholar]
- 35.Sundaresh S, Doolan DL, Hirst S, et al. Identification of humoral immune responses in protein microarrays using DNA microarray data analysis techniques. Bioinformatics 2006; 22:1760–6. [DOI] [PubMed] [Google Scholar]
- 36.Pacheco MA, Elango AP, Rahman AA, Fisher D, Barnwell JW, Escalante AA. Evidence of purifying selection on merozoite surface protein 8 (MSP8) and 10 (MSP10) in Plasmodium spp. Infect Genet Evol 2012; 12:978–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barr PJ, Green KM, Gibson HL, Bathurst IC, Quakyi IA, Kaslow DC. Recombinant Pfs25 protein of Plasmodium falciparum elicits malaria transmission-blocking immunity in experimental animals. J Exp Med 1991; 174:1203–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaslow DC, Quakyi IA, Syin C, et al. A vaccine candidate from the sexual stage of human malaria that contains EGF-like domains. Nature 1988; 333:74–6. [DOI] [PubMed] [Google Scholar]
- 39.Williamson KC. Pfs230: from malaria transmission-blocking vaccine candidate toward function. Parasite Immunol 2003; 25:351–9. [DOI] [PubMed] [Google Scholar]
- 40.van Dijk MR, Janse CJ, Thompson J, et al. A central role for P48/45 in malaria parasite male gamete fertility. Cell 2001; 104:153–64. [DOI] [PubMed] [Google Scholar]
- 41.Drew DR, Sanders PR, Crabb BS. Plasmodium falciparum merozoite surface protein 8 is a ring-stage membrane protein that localizes to the parasitophorous vacuole of infected erythrocytes. Infect Immun 2005; 73:3912–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torres KJ, Clark EH, Hernandez JN, Soto-Cornejo KE, Gamboa D, Branch OH. Antibody response dynamics to the Plasmodium falciparum conserved vaccine candidate antigen, merozoite surface protein-1 C-terminal 19kD (MSP1–19kD), in Peruvians exposed to hypoendemic malaria transmission. Malar J 2008; 7:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Villasis E, Lopez-Perez M, Torres K, et al. Anti-Plasmodium falciparum invasion ligand antibodies in a low malaria transmission region, Loreto, Peru. Malar J 2012; 11:361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.