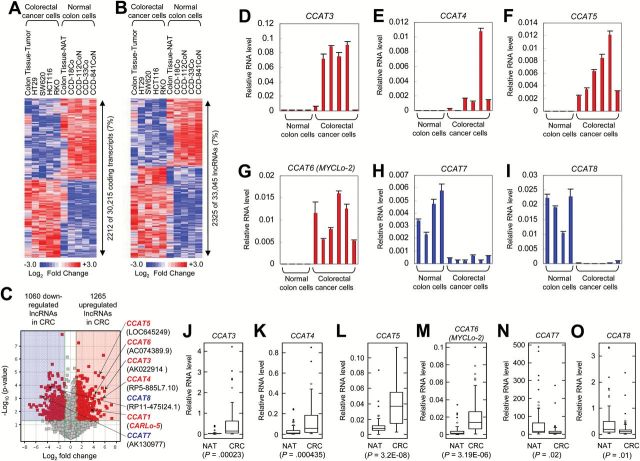
Figure 1.
Identification of long noncoding RNAs (lncRNAs) differentially expressed in colorectal cancer (CRC). A and B) Heatmaps of protein-coding transcripts (A) and lncRNAs (B) dysregulated in CRC (HCT116, RKO, SW620, HT29, and CRC tissues), compared with normal colon (CCD-18Co, CCD-33Co, CCD112CoN, CCD-841-CoN and colon normal adjacent tissues [NATs]). C) Volcano map indicating up- or downregulation of CRC-associated lncRNAs (CCAT1, CCAT3~8). D-G) Verification of induced expression of CCAT3 (D), CCAT4 (E), CCAT5 (F), and CCAT6 (G) in CRC cell lines by using quantitative real-time polymerase chain reaction (qRT-PCR). Data are means ± SD of three independent experiments and each measured in triplicate. H and I) Verification of repressed expression of CCAT7 (H) and CCAT8 (I) in CRC cell lines by using qRT-PCR. Data are means ± SD of three independent experiments and each measured in triplicate. J-O) Differential expression of CCAT3 (J), CCAT4 (K), CCAT5 (L), CCAT6 (M), CCAT7 (N), and CCAT8 (O) in 52 matched CRC and NAT tissues. Shown are box plots with lower and upper bounds of the boxes representing the 25th and 75th quartiles, respectively; whiskers demarcate values within the 10th to 90th percentiles, and solid circles indicate values less than the 10th percentile and greater than the 90th percentile.