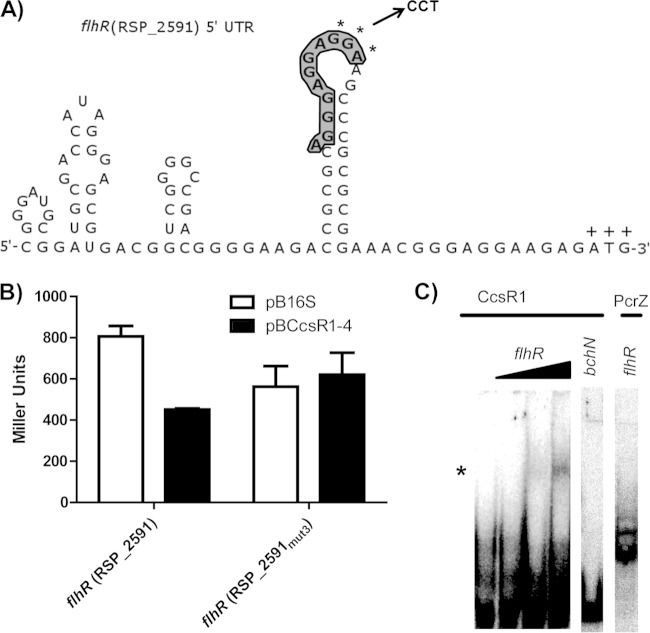
FIG 4.
Effects of mutations in the predicted flhR binding site of CcsR1–4 and interaction of CcsR1 with the flhR mRNA in vitro. (A) The secondary structure of the flhR mRNA displays an exposed binding site for CcsR1 (bold and shaded) that is in relatively close proximity to the ATG codon (marked by plus signs). The bases selected for a triple mutation (GGA) are marked by asterisks, and the corresponding bases introduced for mutation (CCT) are displayed. Secondary structure was predicted by NUPACK (36). (B) A triple mutation within the predicted binding site of the flhR mRNA leads to loss of regulation by CcsR1–4 in the in vivo reporter system. The β-galactosidase activity was measured in cell extracts from R. sphaeroides cultures in the exponential growth phase carrying either pBBR4352 or pBCcsR1–4. (C) CcsR1 interacts with a 177-nt flhR mRNA fragment containing the predicted binging site already at low concentrations in vitro. A 250-fmol sample of CcsR1 was incubated with 25, 250, or 2,500 fmol of the flhR fragment and 2,500 fmol of the bchN control mRNA fragment, while 250 fmol of PcrZ (4) was incubated with 2,500 fmol of the flhR fragment. Changes in gel migration behavior were observable only for CcsR1 with the flhR fragment (marked by an asterisk), while no shift in the gel migration behavior of the controls was observable.