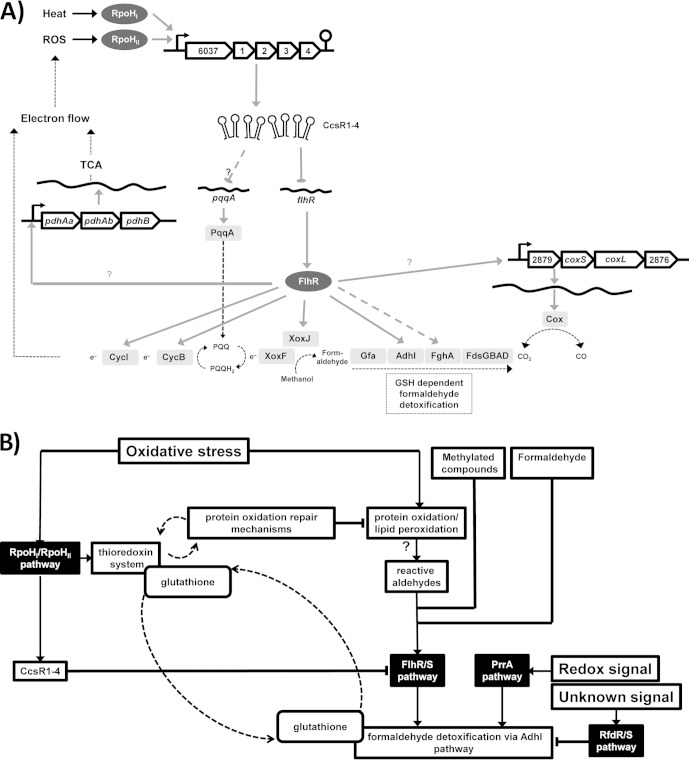
FIG 6.
Metabolic network of CcsR-dependent targets and influence on cell regulatory networks. (A) After stress-dependent transcription, sRNAs CcsR1–4 repress flhR directly and thereby indirectly repress several genes with roles in C1 metabolism and electron transport. Moreover, CcsR1–4 influence pqqA expression through an unknown mechanism. FlhR is a transcriptional activator that has been shown to regulate adhI and cycI in R. sphaeroides (53). cycB and fghA were demonstrated to be regulated by FlhR in P. denitrificans (55), while xoxJ is located in an operon together with cycB in R. sphaeroides. Moreover, xoxJ and cycB also show FlhR dependence in R. sphaeroides and a dependence of the pyruvate dehydrogenase complex gene pdhB on FlhR is indicated. XoxF and XoxJ are potential methanol oxidation proteins. TCA, tricarboxylic acid cycle. (B) Under oxidative stress, thioredoxins and GSH help to prevent oxidative protein and lipid damage by interacting directly with oxidized residues and CcsR1–4 lead to repression of flhR. Through this, the stimulation of the AdhI pathway is indirectly decreased. In addition, the AdhI pathway is known to be regulated by PrrA and RfdR (12, 53). If protein oxidation is not sufficiently prevented, protein carbonylation occurs and reactive aldehydes are produced. These reactive aldehydes probably lead to activation of flhR and more AdhI is produced, which then, with the help of GSH, takes on a putative protective role in the oxidative stress response (54). Through the influence of the CcsR RNAs, fine-tuning of GSH allocation in this network is achieved.