Abstract
Selective pressure to maintain small genome size implies control of transposable elements, and most old classes of retrotransposons are indeed absent from the very compact genome of the tunicate Oikopleura dioica. Nonetheless, two families of retrotransposons are present, including the Tor elements. The gene organization within Tor elements is similar to that of LTR retrotransposons and retroviruses. In addition to gag and pol, many Tor elements carry a third gene encoding viral envelope-like proteins (Env) that may mediate infection. We show that the Tor family contains distinct classes of elements. In some classes, env mRNA is transcribed from the 5′LTR as in retroviruses. In others, env is transcribed from an additional promoter located downstream of the 5′LTR. Tor Env proteins are membrane-associated glycoproteins which exhibit some features of viral membrane fusion proteins. Whereas some elements are expressed in the adult testis, many others are specifically expressed in embryonic somatic cells adjacent to primordial germ cells. Such embryonic expression depends on determinants present in the Tor elements and not on their surrounding genomic environment. Our study shows that unusual modes of transcription and expression close to the germline may contribute to the proliferation of Tor elements.
INTRODUCTION
The multiplication of transposable elements (TEs) creates DNA rearrangements that challenge genome stability. Vertebrate genomes are relatively large and contain numerous TEs. Small genomes, in principle more vulnerable to loss-of function mutations, can eliminate TEs through strong purifying selection (1). In rare cases of ‘domestication’, DNA sequences from TEs are exploited by the host genome (2). Tunicates are the sister group of vertebrates (3). There is growing interest in the larvacean class of tunicates, due to their key roles in marine ecosystems (4), and because some species are amenable to experimental work. Oikopleura dioica is a commonly found larvacean with separate sexes (5). Swimming larvae hatch a few hours after fertilization. After metamorphosis, the juvenile repetitively synthesizes a complex filter-feeding apparatus (the ‘house’) (6). Mature animals release their gametes in the surrounding sea water, completing the short life cycle in 6 days at 15°C. The Oikopleura genome is very small and has undergone dramatic structural changes from the ancestral chordate state (7). It is exceptionally compact and the TE complement has been profoundly remodelled (7,8), to the extent that many families of TEs have disappeared, especially among non-LTR retrotransposons (8). However, the genome contains a new, diversified family of Tor (Ty3/gypsy Oikopleura Retrotransposons) Long Terminal Repeat (LTR) retrotransposons. Even though Tor and gypsy elements are structurally similar, Tor families are fairly divergent from gypsy elements and were found only in Oikopleura (8). By using a partially sequenced genome, we previously observed a high nucleotide identity between different copies, suggesting they correspond to multiple integrations of an active Tor element (8). Several Tor elements possess a third open reading frame (ORF) that may encode an envelope protein (Env) (8) and as such, they could represent endogenous retroviruses (ERVs) (9). Env proteins comprise structurally diverse glycoproteins located on the envelope of various viruses. The common function of Env proteins is to facilitate viral entry by promoting membrane fusion (10).
We studied the expression of Tor elements that carry an env gene, using RACE (Rapid Amplification of cDNA Ends), RT-PCR, whole mount in situ hybridization (WISH) and expression of reporter constructs in early embryos. In known ERVs, transcription usually starts at a promoter located in the 5′LTR and terminates in the 3′LTR. A long genomic RNA (gRNA) is produced, from which alternative splicing produces env mRNA (11–14). Depending on the host species, ERV genes can be expressed in a variety of cells and tissues, either germinal or somatic. For example, Drosophila ERVs expressed in ovarian follicle cells reach the germline via Env-mediated infection (15). Mouse ERVs are expressed in embryonic primordial germ cells (PGCs) thus providing direct access to the germline genome (16). Various mechanisms are employed to defend the germline from ERVs, including small RNA-mediated silencing (17). Our results show that Tor Env is a membrane-bound glycoprotein, similar to Env present in Drosophila ERVs (12,15). In several Tor elements, env expression is driven by an internal promoter and not by the 5′LTR. Such an unusual mechanism could play a role in Tor RNA metabolism. Some Tor elements are transcribed in the adult testis and we show that several others are specifically expressed in the embryo. We observed early embryonic expression in various somatic cells located near PGCs, which may facilitate access of Tor elements to the germline. These results indicate how a family of retrotransposons can proliferate in a highly compact genome.
MATERIALS AND METHODS
Protein sequence analysis
We interrogated the Oikopleura genome (7) to identify genomic scaffolds that matched Tor pol and/or env sequences (8) and mapped the LTRs by aligning their flanking sequences. Some ORFs were extended by correcting sequencing errors after multiple alignment of shotgun reads. Functional predictions of Env were obtained using the Eukaryotic Linear Motif (ELM) resource (18) and the domain annotation of Pol was based on the Pfam database. We aligned Pol sequences that encompass reverse transcriptase and integrase domains using MUSCLE (19) and GBLOCKS (20). Gaps were removed from the alignments and phylogenies built with PhyML, applying default parameters and the aLRT statistical test. We used reciprocal BLAST and multiple sequence alignment to annotate Oikopleura piwi and ago genes. The phylogeny of candidate Argonaute proteins was based on PAZ and PIWI domains.
Animal breeding
Oikopleura dioica collected from fjords around Bergen were cultured and bred as described (5). To preserve sperm samples from individual males, we followed a new cryopreservation procedure (Bouquet et al., in preparation). Synchronous embryos were obtained by in vitro fertilization in artificial, filtered sea water (AFSW, Red Sea, 30.1–30.5% salinity) and left to develop at 20°C.
Cloning
For the synthesis of RNA probes (Supplementary Figure S1), we amplified gene fragments with polymerase chain reaction (PCR) using specific primers and cDNA libraries from mixed developmental stages (1–6 h pf). The PCR products were cloned in pCRII-TOPO (Invitrogen) or pGEM-T vectors (Promega), prior to linearization for in vitro transcription with T7 or SP6 RNA polymerases (Roche). For expression in HEK293T cells, we cloned the complete env ORFs in C-terminal fusions with V5/6His tag in pCDNA 3.1-TOPO vector (Invitrogen). To construct pCTor3b-2 and pCTor4b-1, we generated DNA fragments containing part of env, fused with the V5/6His tag, followed by a stop codon and a restriction site (Figure 6A). These were then digested with the appropriate enzymes and ligated to upstream and downstream DNA fragments in order to reconstruct a Tor element that carried the full gag to the 3′LTR sequence. The resulting inserts were PCR amplified and cloned into the pCRII-TOPO vector.
Figure 6.
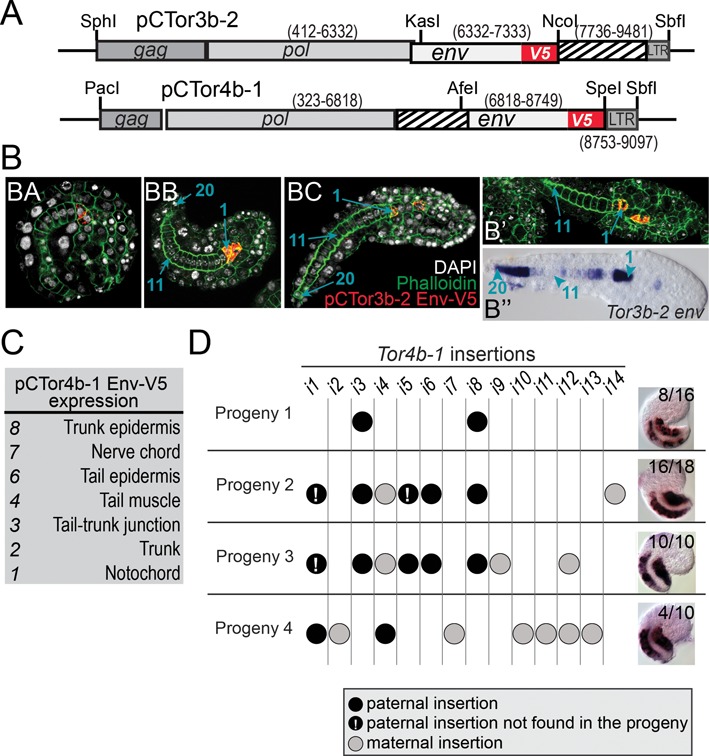
Autonomous expression of Torgenes. (A) Schematic representations of the expression constructs tested in Oikopleuraembryos. The numbers indicate coordinates on TorDNA, striped boxes represent non-coding sequences. (B) pCTor3b-2 drives Env expression in the anterior cells of the notochord. (BA), embryo before hatching; (BB) and (BC), embryos after hatching showing a complete notochord with 20 cells (blue arrows); (B’) and (B’’)), comparison of pCTor3b-2 activity with the expression pattern of Tor3b-2 env in wild-type embryos. (C) pCTor4b-1 expresses Env in various tissues. The table indicates the number of positive embryos showing expression in the same tissue (Supplementary Figure S7A). (D) Tor4b-1 copies and their envexpression pattern. The table shows the presence of insertions i1 to i14 in F1 animals from individual crosses. Segregation in the progeny was followed using Southern blotting (Supplementary Figure S7B). On the right, micrographs show WISH patterns obtained in the progeny using the antisense Tor4b-1 env probe. Numbers indicate signal frequency.
RNA profiling
Large (>200 nt) total RNA was extracted from samples with the mirVana miRNA isolation kit (Ambion), following manufacturer's recommendations. After DNase treatment, RNA was purified with organic extraction and ethanol precipitation. The mapping of RNA 5′ ends was performed with a protocol adapted from the FirstChoice RLM-RACE kit (Ambion) or alternatively, with the SMARTer PCR cDNA synthesis kit (Clontech). To prepare cDNA for expression profiling, we annealed 50 or 100 μg.ml−1 total RNA for 3 min at 65°C in the presence of either 20 μg.ml−1 Oligo(dT)12–18 (Invitrogen), 2 μM random 10-mer (Ambion) or 200 nM gene-specific primer. The mix was cooled to 50°C for 1 min in RT buffer (50 mM Tris–Cl, 75 mM KCl, 3 mM MgCl2, pH 8.3) and supplemented with 10 mM dithiothreitol (DTT) and 1 mM dNTP, prior to addition of 0.025 U.μl−1 of Superscript III RT (Invitrogen). The reactions were run for 90 min at 50°C, RT was inactivated 15 min at 70°C and tubes were put on ice prior to treatment with.0.05 U.μl−1 of RNase H (Invitrogen) for 30 min at 37°C. For PCR amplification with specific primers, we used Advantage 2 DNA polymerase (Clontech) in a two-step protocol that includes an annealing/elongation step at 68°C and a moderate number of cycles (up to 31).
Cell culture and biochemical assays
HEK293T cells were grown in standard conditions. Cultures in 12-well plates were transfected with Polyfect (Qiagen) using 0.5 μg of pCDNA-Tor Env following manufacturer's recommendations. To determine subcellular localization, cells were washed with phosphate-buffered saline (PBS) 48 h after transfection and lysed by 10 passages through a 23G needle in the presence of subcellular fractionation buffer (SFB; 0.25 M sucrose, 20 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 1 mM ethylenediaminetetraacetic acid (EDTA), 1 mM ethylene glycol tetraacetic acid (EGTA), 1 mM DTT, pH 7.4) supplemented with Complete (Roche). After 20 min on ice, the lysate was clarified for 10 min at 10 000 g, and the supernatant was centrifuged 1 h at 115 000 g in a SW55Ti rotor. The cytoplasmic supernatant was concentrated with Centriprep YM-50 (Millipore), resuspended in Radioimmunoprecipitation assay buffer (RIPA; 50 mM Tris–Cl, 0.15 M NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulphate (SDS), 1 mM EDTA, pH 7.5) and flash-frozen in liquid N2. The membrane pellet was washed with SFB, centrifuged and resuspended in RIPA prior to flash-freezing. Proteins in cytoplasmic and membrane fractions were quantified with the bicinchoninic acid (BCA) protein assay kit (Pierce) prior to western blotting. Two microgram aliquots of each fraction were run on a 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis, and transferred to a polyvinyl difluoride (PVDF) membrane (Immobilon-P, Millipore). The presence of tagged Env, β-Tubulin and Cadherin was detected with HRP-conjugated antibodies (Anti-V5 (Invitrogen), Anti-beta Tubulin and Anti-pan Cadherin (Abcam), respectively). Chemiluminescence (Amersham ECL prime, GE) was detected using Chemidoc XRS+ (Biorad). To test for protein glycosylation, total cell extracts were treated with Protein Deglycosylation Mix (NEB) prior to western blotting.
RNA in situ hybridization
Animals were fixed in PAF (4% paraformaldehyde, 0.1 M MOPS, 0.5 M NaCl, pH 7.5) overnight at 4°C, washed with PBSTX (PBS, 0.1% Triton X-100) and 50 mM Tris–Cl (pH 8.0) before treatment for 3 min with 10 μg.ml−1 of proteinase K at 37°C. Permeabilized samples were fixed in PAF for 20 min at room temperature, washed with PBSTX and prehybridized in HB buffer (50% formamide, 5× saline-sodium citrate (SSC) buffer, 0.1% Triton X-100, 50 μg.ml−1 tRNA, 50 μg.ml−1 heparin). Antisense RNA probes labelled with digoxigenin or fluorescein (21) were denatured 2 min at 90°C in HB and incubated with samples overnight at 60°C. Following washes, samples were treated 30 min at 37°C with 20 μg.ml−1 RNase A in TNE buffer (10 mM Tris–Cl, 0.5 M NaCl, 1 mM EDTA, pH 7.5). Reactions were stopped by incubation for 30 min at 60°C in TNE, prior to blocking (PBSTX with 1% Blocking Reagent (Roche) and 1% lamb serum) for 1 h at room temperature. Samples were then incubated overnight at 4°C, with Anti-Digoxigenin-AP (Roche) diluted 1/3000. Detection with NBT/BCIP was performed as described by Seo et al. (22). For double fluorescent WISH, samples were quenched with 0.3% hydrogen peroxide in PBSTX for 30 min at room temperature prior to antibody blocking steps. Anti-Fluorescein-POD and Anti-Digoxigenin-POD (Roche) were used in blocking solution at 1/1000 and 1/300 dilutions respectively and samples were stained for at least 90 min using the Tyramide Signal Amplification (TSA) plus kit (Perkin Elmer).
Southern and northern blots
For Southern blots with Oikopleura genomic DNA, we chose an appropriate combination of probe and restriction enzyme that would result in a single band for each Tor copy. The genomic DNA from single individuals was extracted using a standard protocol with guanidium thiocyanate (GTC) extraction, followed by proteinase K and RNase A treatment. Purified DNA was digested with SacI (NEB) overnight at 37°C, precipitated with ethanol and run overnight on a 0.7% agarose-TAE gel in the presence of ethidium bromide. The gel was checked under UV illumination for DNA digestion, then depurinated for 5 min at room temperature in 0.25 M HCl. DNA was denatured in gel prior to downward capillary transfer in 10× saline-sodium phosphate EDTA buffer (SSPE) and crosslinking to a nylon membrane (Hybond N+, GE). For northern blots, 0.25 μg of poly-A+ RNA (Dynabeads mRNA direct, Ambion) were run on 0.8% agarose-formaldehyde gels. Following downward capillary transfer in 5× SSPE and 10 mM NaOH, RNA was crosslinked to Hybond N+. Membrane hybridization was performed overnight with 1 × 106 cpm of denatured probe, in 12 ml Ultrahyb (Ambion); at 68°C for northern blots or at 50°C for Southern blots. Following extensive washing with SSPE and SDS at 65°C, the blotted membranes were exposed to a phosphorimaging plate for one week. The signal was recorded on a Typhoon FLA 9500 scanner, set at 1000 V sensitivity and 25 μm pixel size.
Expression of recombinant Tor Env in embryos
Oocytes collected from individual O. dioica females were fertilized with pooled sperm for two min in AFSW at 18°C. After transfer in fresh AFSW supplemented with 1 mM CaCl2 (AFSW-Ca2+), the egg chorion was permeabilized at 5 min pf by a 30 s treatment with 0.04% pronase. Eggs were washed with AFSW-Ca2+ and transferred to the micro-injection dish at 8 min pf. The injection mix (15 ng.μl−1 circular plasmid DNA, 20 mM Alexa Fluor 633) was delivered using a laser-pulled quartz capillary with filament (Sutter Instruments), connected to a FemtoJet (Biorad). Eggs were injected until the first cleavage and then transferred to fresh AFSW. The resulting embryos or larvae were fixed in ice-cold PAF for 1–3 h, then washed extensively in PBSTE (PBS, 0.1% tween-20, 1mM EDTA) in the presence of 0.1 M glycine and incubated 1 h at room temperature in blocking solution (10% heat-inactivated FCS, 3% BSA, 0.02% sodium azide in PBSTE). Samples were further incubated 4 days at 4°C in blocking solution supplemented with 1/500 Anti-V5-Cy3 (Sigma), prior to 30 min counterstaining at room temperature with 2% Alexa Fluor 488 Phalloidin (Molecular Probes). After five washes with PBSTE in the presence of 1% BSA, samples were mounted in Slowfade Gold with DAPI (Molecular Probes) and observed with a Leica TCS laser scanning confocal microscope.
Analysis of the Oikopleura transcriptome
We used the OikoBase server (23) to reveal cDNA hybridization intensities on genome tiling arrays at different developmental stages (not including mature females). We analysed 100 Tor sequences, which included complete elements and fragments containing pol and/or env ORFs found in the genomic reference assembly. Tiling array data analysis was performed with UPGMA clustering.
RESULTS
Tor elements encode viral-like transmembrane glycoproteins
Classification of Tor elements
Using a fully assembled genome sequence representing two distinct haplotypes (7), we detected Tor sequences in 167 genomic scaffolds and identified 38 elements carrying a candidate env gene. Phylogenetic analyses of Pol using Maximum-Likelihood or Neighbour-Joining methods, reproducibly revealed Tor1, Tor2, Tor3a, Tor3b, Tor4a and Tor4b groups (Figure 1A). Their positions relative to other gypsy elements were variable and weakly supported. In Tor4b elements, we systematically found a space between pol and env genes (Figure 2A). Half of the Tor elements with env belong to Tor3a, with the others belonging to Tor3b and 4b. A few Tor elements without env were also found in these groups.
Figure 1.
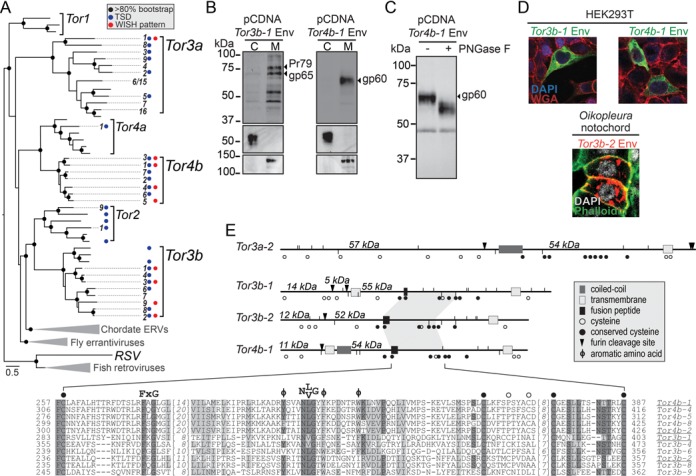
Classification and biochemical features of Tor envelope proteins. (A) Phylogeny of Tor elements based on Pol. Inside each group, numbered branches indicate elements whose embryonic expression was tested with WISH. The elements Tor3a-6 and -15 have very similar Pol but other genes are divergent. Red dots indicate elements showing tissue-specific WISH patterns in the embryo. Blue dots show when Target Site Duplications are present. Scale bar shows number of substitution per site. RSV, Rous Sarcoma Virus. (B) Subcellular fractionation of Tor3b-1 and 4b-1 Env. Cytoplasm (C) and membranes (M) fractions were purified from HEK293T cells expressing tagged Env and analysed by western blot. The top panel shows detection of glycosylated Env peptide (gp) and Env precursor before furin cleavage (Pr). The fusion tag used in these experiments adds an extra 5 kDa to the protein molecular weight (MW). Middle and bottom panels show detection of β-tubulin and Cadherin, respectively. (C) Deglycosylation assay of Tor4b-1 Env. Treatment of cell extracts with Peptide-N-Glycosidase F resulted in an Env bandshift. (D) Micrographs showing Tor3b-1 and 4b-1 Env localization in human and Oikopleura cells. Nuclei were stained with DAPI and cell boundaries were stained using Wheat Germ Agglutinin or Phalloidin. (E) Primary structure of Tor Env. At top, schematic representation of Env from selected Tor3a, 3b and 4b elements. The arrows show predictions of furin proteolytic cleavage and numbers in italic indicate the MW of the resulting fragments. Vertical lines show residues favourable for N-glycosylation (upwards) and glycosaminoglycan attachment (downwards). The shaded area shows the region with highest conservation found in Tor4/3b Env, corresponding to the multiple sequence alignment below. Positions highlighted in dark grey show amino acids identical in at least half of Tor3b and 4b sequences, positions highlighted in light grey show conservative substitutions. Full-length env cDNAs were obtained for elements whose names are underlined.
Figure 2.
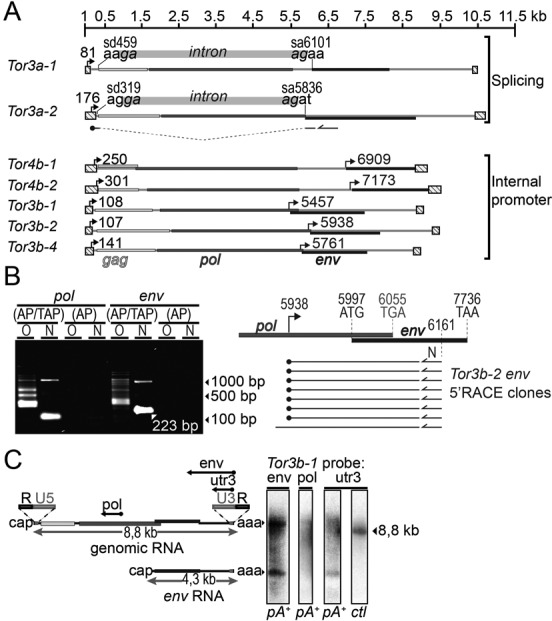
Characterization of Tor TSSs. (A) Genetic organization of Tor elements. LTRs are shown as striped boxes, arrows and numbers indicate Transcription Start Sites (TSSs) mapped with 5′RACE. Intron positions and border sequences are shown for Tor3a elements. sd, splice donor site; sa, splice acceptor site. The structure of a 5′RACE product obtained with an env-specific primer (half arrow) is shown for Tor3a-2. (B) Representative 5′ RLM-RACE (RNA Ligase-Mediated RACE) experiment for the mapping of TSSs of Tor3b-2. On the left, the gel shows products from first and second rounds of PCR with outer (O) and nested (N) primers. Treatment of RNA prior to cDNA synthesis: AP/TAP, dephosphorylation followed by Tobacco Acid Pyrophosphatase; AP, dephosphorylation only. The white arrow indicates the major env RACE product and its size. On the right, eight RACE clones obtained from env mRNA (Supplementary Figure S3) are represented, along with the predicted translation initiation and termination codons for pol and env. (C) Northern blot analysis shows gRNA and env mRNA. Hybridization was performed using three different antisense probes, either on mRNA extracted from embryos (pA+), or on in vitro synthesized Tor3b-1 RNA (ctl). R, sequence repeated in the 5′ and 3′; U5 and U3, unique sequence in the 5′ and 3′, respectively.
Biochemical study of Tor Env
The comparison of Env proteins from Tor3b and 4b revealed highly conserved residues, forming motifs absent in Tor3a. These conserved features, found upstream of a C-terminal transmembrane domain, include a putative fusion tripeptide F-X-G (24), a N-[L/V]-G motif flanked by aromatic residues and a set of seven cysteines (Figure 1E and Supplementary Figure S2). Using human HEK293T cells, we expressed Tor3b-1 and 4b-1 Env proteins fused to a C-terminal tag. Env was associated with cell membranes during subcellular fractionation (Figure 1B). Immunostaining of transfected cells did not allow conclusive identification of the organelles where these Env proteins were located. We also expressed Tor3b-2 Env in Oikopleura embryos by micro-injection of a reporter construct (see ‘Tissue-specific expression of Tor is driven by internal regulatory sequences’ of ‘Results’ section). In this case, we observed Env localized on the plasma membrane and in perinuclear structures, most likely the endoplasmic reticulum (ER) (Figure 1D). The presence of glycosylated residues on Tor4b-1 Env expressed in human cells also indicated ER addressing (Figure 1C). Oligosaccharides in such residues can participate in the fusion of viral envelopes with cell membranes (25). Finally, we identified Env-derived peptides (gp65 for Tor3b-1 and gp60 for Tor4b-1) whose sizes were in agreement with products of furin-mediated cleavage, a step required for viral envelopes to become fusion-competent (10). Taken together, these experiments characterize Tor Env as a transmembrane glycoprotein that may be involved in intercellular transfer of virus-like particles (VLPs).
The synthesis of env RNA begins either in the 5′LTR or at an internal promoter
We used a 5′RACE approach to map transcription start sites (TSSs). Using gag or pol primers, we found a TSS within the 5′LTR for each element tested (Figure 2A). Using env primers for Tor3a-1 and -2, we found that env mRNA was spliced and transcribed from the 5′LTR as well (Supplementary Figure S3). We mapped splice donor sites close to the beginning of gag and splice acceptor sites upstream of env. In both cases the intron had a non-canonical donor site GA, but an acceptor site similar to those of GT-AG introns in the genome (7). More surprisingly, env-primed 5′RACE products from Tor3b and 4b elements were systematically devoid of LTR sequences and lacked evidence of splicing (Figure 2B). In five cases, the cDNA matched a sequence beginning upstream of env, either in pol for Tor3b elements or in the intergenic space that follows pol in Tor4b elements. Since our cloning protocol selects for capped RNA, these RACE products most likely represent transcripts synthesized from an internal promoter downstream of the 5′LTR. We refer to this promoter as Penv, and to the LTR promoter as Pltr. We further characterized env mRNA with northern blotting using antisense probes that cover pol, env or the 3′UTR of Tor3b-1 (Figure 2C). We identified two transcripts expressed in the embryo. The longer one corresponds to the gRNA and the shorter one to env mRNA. Further RACE analyses showed that 5′ and 3′ ends of Tor3b-1 gRNA carry R/U5 and U3/R arrangements, respectively, which are required for conservative replication (26).
Insertion polymorphism and developmentally-regulated expression
Detection of recent integration
We characterized 32 distinct elements from the sequenced genome. Each element is represented by at least one full-length copy displaying hallmarks of recent integration. These include nearly-identical LTRs (98.5% on average), TSDs (Target Site Duplications, 4 or 5 bases) and intact ORFs (Supplementary Table S1). We investigated additional indications of activity by analysing insertion polymorphisms of Tor3b-1, 3b-2 and 4b-1 (27). Most insertions were hemizygous, i.e. present in one haplotype of the genome and absent in the other (Supplementary Figure S4). Genotyping of wild or cultured animals showed that genomic insertions were hemizygous and present in a small minority of individuals (<5%). Southern blotting analyses of a given element in sperm DNA from different individuals revealed a small number of insertions in each individual and variable patterns of insertions among individuals (Figure 3A). Such polymorphism indicates that the reference genome contains only a few of the full complement of existing insertions. Several insertions absent from the genome assembly were indeed identified either with the transposon display technique (28) or by cloning chimeric transcripts that included Tor and flanking sequences (Figure 3B, Supplementary Table S2). Taken together, these results indicate that some Tor elements are active and have recently integrated copies in Oikopleura germline DNA.
Figure 3.
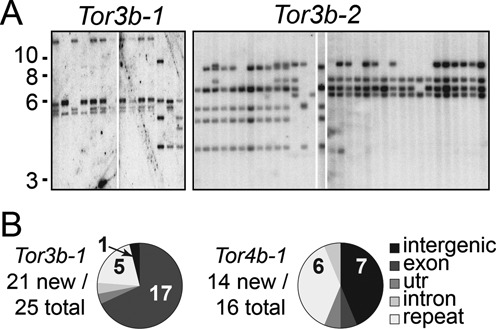
Tor insertion polymorphism. (A) Southern blot analysis showing single copies of Tor. Lanes were loaded with genomic DNA from individual males. Sizes are indicated in kilobases (kb). (B) Characterization of Tor insertions. New Tor insertions, absent from the genome assembly, were identified in randomly sampled DNA. Pie charts show the proportion of insertion sites present in different contexts.
Spatio-temporal analysis of Tor expression during embryogenesis
To better understand the mechanisms by which Tor elements proliferate, we studied the expression of pol and env during development. We first tested for the presence of transcripts in cDNA samples prepared at successive developmental stages using RT-PCR and hybridizations on genome tiling arrays (23) (Figure 4B and Supplementary Figure S5C). Both techniques showed that for most elements, Tor RNA seemed to be absent or present in very low amounts during the first hours of development. Embryonic expression of Tor was first detected between 80 and 100 min pf, during early gastrulation (29) (Figure 4BB). On tiling arrays, we found that 29 out of 100 elements showed significant expression during tailbud and hatching stages (Figure 4BA). Among these, 13 elements were expressed only during embryogenesis, with the highest levels at tailbud or hatching stages. Other elements were expressed mainly after the larval tailshift. RT-PCR assays were in agreement with the tiling array data and for three Tor3b/4b elements we also confirmed the presence of gRNA and env mRNA in embryos (Figure 4BC). Expression profiling showed that for most Tor elements, increases in gene expression occurred during specific developmental stages. We next checked if Tor RNA is present in somatic tissues or in the germ line.
Figure 4.
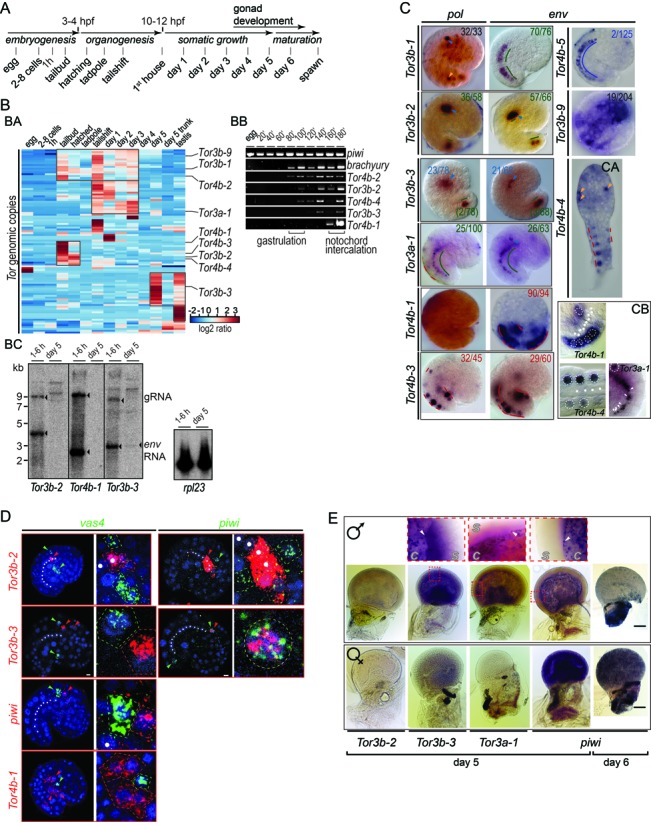
Expression of Tor during development. (A) Schematic representation of the Oikopleura life cycle. (B) Tor expression in Oikopleura cDNA collected at different stages. (BA) The heat map represents variation of Tor expression as shown by cDNA hybridization on genomic tiling arrays. Hierarchical clustering reveals five groups of elements with similar expression, highlighted with squares on the heat map. (BB) RT-PCR profiling of piwi, brachyury and Tor env within 3 h pf. (BC) Northern blots performed with antisense env probes show Tor expression in poly-A+ RNA extracted either from embryos or from day 5 animals. The housekeeping gene rpl23 was used as a loading control. (C) WISH samples stained with NBT/BCIP show gene expression patterns obtained with antisense pol and env probes. Animals were collected at the tailbud stage or after hatching (CA). Yellow and blue arrows mark uncharacterized cells present in the epiderm and mesenchyme, respectively. Lines highlight positive cells in the notochord (green), tail muscle (red) and tail nerve chord (blue). The proportion of samples showing the same pattern is indicated. (CB) shows a magnified view of env signal in the tail. Dotted circles, muscle cell nuclei; white dots and arrows, notochord cell nuclei. (D) Double fluorescent WISH showing expression of Tor, vas4 and piwi. Samples were collected at 160 min pf for Tor4b-1 and at 180 min pf for the other probes. Probes were labelled with FITC (green) or Cy5 (red) and nuclei were stained with DAPI (blue). Arrows mark positive cells. Magnifications show signals in neighbouring cells or in the same cell for Tor3b-3 (yellow dotted circle). Scale bar, 5 μm. (E) WISH samples showing expression of piwi and env in day 5 and 6 animals: males (top), females (bottom). Magnification of the testis (top row) reveals follicular cells (arrows) present in day 5 animals at the interface between the gonad cavity (c) and the syncytium (s). Scale bar, 200 μm.
For 27 Tor elements, we examined the expression of pol and env RNA in embryos with WISH. Using antisense probes, we revealed tissue-specificity for nine elements (Figure 4C and Supplementary Figure S5E). Expression patterns of pol and env were identical for Tor3a-1, where a single Pltr was mapped. When both Pltr and Penv were present, two different patterns were observed. Identical pol and env expression patterns of Tor3b-2 suggest a common set of regulatory sequences acting on both promoters (Supplementary Figure S5A and B). Conversely, the distinct patterns seen for Tor3b-1 and 4b-1 suggest promoter-specific regulation.
Five elements exhibited strong, reproducible env expression in the notochord or in muscle. Tor4b-1, -3 and -4 were expressed with the same intensity in almost every cell of the tail muscle. Tor3b-1 and -2 were expressed in most notochord cells but with more variable intensity. Tor3b-2 expression in anterior cells appeared after 2 h of WISH staining while expression in posterior cells was detected only after overnight staining. In contrast, Tor3b-3 rarely showed expression in the notochord and was more often expressed in the dorsal trunk. Similar to Tor4b-3 and -4, Tor3a-1 RNA was detected mostly in the nucleus (Figure 4CB), showing possible sequestration of immature transcripts. For most elements tested, minor expression was often observed in isolated cells of the trunk and epidermis. Finally, Tor3b-9 and Tor4b-5 showed tissue-specific expression in a small minority of embryos, possibly because of a copy-specific activity dependent on external, host genome cis-regulatory sequences. These results show that several Tor elements are somatically expressed during embryogenesis. Hybridizations on genome tiling arrays revealed increased expression levels for a subset of elements at this stage (Figure 4BA). Therefore, somatic expression likely reflects specific activation and not genome-wide de-repression of Tor promoters (30). In Oikopleura embryos, notochord and tail muscle cells are fate-restricted as early as the sixth cleavage (29). Tor RNA is thus present when the embryo has a small number of cells.
Tor is expressed in somatic cells close to the PGCs
The germ and stem cell markers vasa and piwi (31) participate in the silencing of TEs (32,33). Comparison of Argonaute protein sequences in the Oikopleura genome identified a single Piwi, sharing diagnostic residues with fly and vertebrate Piwi (Supplementary Figure S6). Phylogenetic analysis suggests that Oikopleura Piwi is a divergent orthologue of one of the two Piwi paralogues usually found in metazoans (Figure 5). Four vasa genes are present in the Oikopleura genome and we focused on vas4 as it is the only Vasa paralogue expressed in PGCs (Lisbeth Charlotte Olsen, in preparation).
Figure 5.
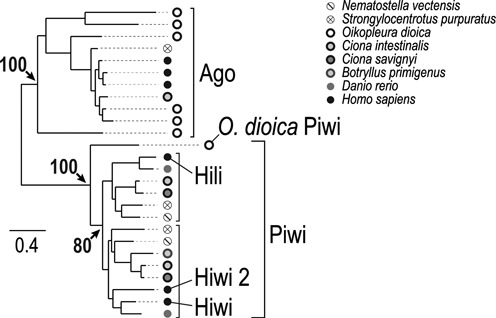
Phylogeny of Oikopleura Argonaute proteins. The phylogenetic tree shows that several Oikopleura proteins belong to the Ago subfamily, whereas only one protein is present in the Piwi subfamily. Arrows indicate bootstrap values. Scale bar shows number of substitution per site.
Using double fluorescent WISH, we compared the expression sites of Tor with those of vas4 and piwi. In tailbud embryos, vas4 and piwi were expressed separately in distinct, adjacent cells of the dorsal trunk (Figure 4D). Tor3b-3 was expressed in one piwi-positive cell. At 160 min pf, muscle cells expressing Tor4b-1 surrounded vas4-positive cells. Then at 180 min pf, vas4-positive cells came into contact with anterior notochord cells expressing Tor3b-2. In day 5 animals, piwi was expressed in the immature gonads of both sexes (Figure 4E). At day 6, piwi was expressed only in females. We observed a strong signal in day 5 testes for Tor3b-3 and 3a-1, whereas in the ovary there was either no signal, or a diffuse signal for Tor3b-3. Tor and piwi expression in the testis was restricted to a cell layer covering the internal surface of the gonad cavity, previously identified as follicle cells (34). Our results show that expression of Tor in somatic tissues was located in proximity to germ cells. Tor RNA seemed mostly absent from germ cells themselves.
Tissue-specific expression of Tor is driven by internal regulatory sequences
Tor elements may carry regulatory sequences driving the tissue-specific expression of pol and env in the embryo. Alternatively, such expression could be influenced by external regulatory elements near their integration sites. We designed reporter constructs to test the expression of Tor3b-2 and 4b-1 in vivo. We included the full DNA sequence of each element minus the 5′LTR and fused an epitope tag to Env (Figure 6A). The resulting plasmids, pCTor3b-2 and pCTor4b-1, were injected as circular DNA into fertilized eggs prior to the first cleavage. Env expression, assayed by immunostaining, was detected in ∼10% of embryos that survived the injection. Based on previous experiments, the injected material is most likely maintained out of the chromosomes. pCTor3b-2 was always expressed in the anterior-most notochord cell and often in a single cell located next to it (9 of 22 samples) (Figure 6B). The expression of pCTor3b-2 was not detected in the central and posterior notochord (Figure 6B’ and 6B’’). We previously noted that native expression of Tor3b-2 was indeed much stronger in the anterior notochord.
In contrast to our observations with pCTor3b-2, the expression of pCTor4b-1 was variable and did not reproduce the muscle-specific pattern of Tor4b-1 (Figure 6C and Supplementary Figure S7A). At least two interpretations may reconcile the variable expression of pCTor4b-1 with the native expression of Tor4b-1 in muscle. First, the construct may lack repressive elements that normally restrict expression to tail muscle. For instance, binding of repressors to the 5′LTR can lead to proviral silencing in embryonic cells (35). Second, muscle-specific expression could require external regulatory elements that normally act on some Tor4b-1 insertions but not on the injected construct. To test this latter hypothesis, we checked if variable integration sites might affect expression of Tor4b-1 in muscle. For this, we produced several families from different parents, in which Tor4b-1 genotyping and WISH were combined. Genotyping was restricted to male offspring, which yield sufficient amounts of DNA. In each F1, most Tor4b-1 copies present in fathers were also detected in their sons (Figure 6D and Supplementary Figure S7B). Overall, the results indicate that expression of Tor4b-1 in muscle was not due to one specific insertion of the element (compare for example crosses 1 and 4, in Figure 6D). Therefore, the muscle-specific expression is probably driven by internal regulators present in Tor4b-1 but omitted in the pCTor4b-1 construct.
DISCUSSION
Our study supports ongoing activity of Tor elements, in providing evidence of recent integrations, autonomous tissue-specific expression and a potential role of Env in cell-to-cell transfer. Tor polymorphism suggests turnover with strong purifying selection (1,36). No homozygous insertions have been detected thus far. Overall, the genomic features of Tor elements are reminiscent of TE families described in other small genomes, for which recent activity is either proposed or well documented (1,37,38). The preservation of env genes in a normal retroviral arrangement suggests a function in the lifecycle of Tor and not their domestication by the host (2). We show here marked differences between env genes from group Tor3a and those from groups Tor3b/4b. A common ancestral gene may have undergone rapid divergent evolution, perhaps due to coevolution of Env proteins and their receptors (39). As reported for some fly and nematode ERVs (9), Tor ancestors could also have acquired env from a variety of exogenous viruses during evolution. The major difference in the mode of env expression reported here, with or without internal promoters, could indicate that Tor3a and Tor3b/4b originate from two divergent, independently endogenized, retroviruses.
Acquisition of a new internal promoter for env expression
In complex retroviruses, some accessory genes are transcribed from an internal promoter located outside the LTR (40,41). However, to the best of our knowledge, alternative splicing of the gRNA is the only mechanism reported for env expression in retroviruses (12–14). Our 5′ RACE and reporter expression results show that in some Tor elements env is, instead, expressed independently from the LTR. The purpose of this novel mechanism remains unknown, and could contribute to fine-tune levels of gRNA and env RNA. Indeed, splicing must be tightly regulated in order to retain some gRNA for gag-pol translation and encapsidation (13). The presence of Penv renders the production of Env independent of gRNA splicing in some Tor3b/4b elements. For these elements, splicing could be strongly inhibited by signals such as exonic splicing silencers found in retroviruses (42). In the absence of splicing, the availability of gRNA for VLP assembly would depend solely on the output of Pltr. The acquisition of Penv may also have facilitated env expression, as Tor3b/4b mRNA was easily detected by northern blots. In contrast, the Tor3a elements studied so far seem unlikely to yield significant amounts of functional transcripts. Further insight could be obtained by testing whether splicing contributes to the silencing of Tor3a (43,44).
Somatic expression of Tor could promote germline infection
The expression of TEs in the gonads promotes access to germline DNA, as documented for the fly ovary and mouse testis (12,45). Tor RNA is expressed in follicle cells of the Oikopleura testis and we cannot exclude that low amounts of transcripts are present in developing sperm cells as well. The expression of TEs in animal embryos has been frequently observed (16,46–48), but the mechanisms permitting such expression are not well documented.
A number of studies have shown that Piwi and Vasa can participate in a complex mechanism that represses TEs (33,49). Our results show distinct expression patterns for vas4 and piwi in Oikopleura embryos, suggesting that they play separate roles at this stage. Supporting this idea, piwi restricts vasa expression in the sea urchin embryo (50) and expression in distinct cells was also shown in the fly embryo. In the latter case, piwi and several TEs are expressed in somatic cells of the embryonic gonad, whereas adjacent germ line cells express vasa (51). We also showed co-expression of piwi and Tor3b-3 in the same cell, whereas we did not yet observe co-expression of vas4 and Tor. As proposed for Drosophila (51), piwi-expressing cells may participate in germ line development and could initiate silencing of TEs in Oikopleura embryos. Other Tor elements were strongly expressed in tissues such as the notochord and the muscle, where communication with germ cells is doubtful. Transcription in cells where silencing is less stringent than in germ cells (52) may confer advantages to Tor elements. In this respect, it would be interesting to assess whether or not TE-derived small RNAs and their protein partners are present in the notochord and/or muscle.
A cell lineage study has shown that Tor-expressing notochord and tail muscle cells together account for 17% of the total number of cells in the tailbud embryo at 150 and 180 min pf (29), the stages when we observed a broad activation of Tor. This proportion is probably double in the posterior part of the embryo, where PGCs are located at the early tailbud stage (29). The expression of potentially infectious Tor elements in cells surrounding PGCs is compatible with a possible contribution of somatic expression towards germline transmission of TEs. Tor-derived VLPs produced in somatic cells adjacent to quiescent PGCs in tailbud embryos, could permit the integration of newly synthesized proviruses into germline DNA (Figure 7). These events would occur 1 h before PGCs resume their migration and start to divide. Thorough functional studies will be required to test this scenario (53). A prediction is that somatic expression would only benefit Tor elements having an env gene. Interestingly, of three Tor3b/4b elements without env that were tested thus far, none of them is tissue-specifically expressed in the embryo.
Figure 7.
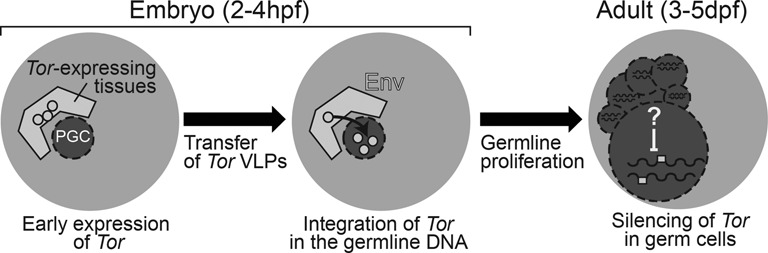
Hypothetical germline infection by Tor elements. Left: during embryogenesis, Tor transcripts are produced in somatic tissues adjacent to PGCs and their translation gives rise to VLPs (circles). Middle: Env protein could play a role in the infectious transfer of VLPs to PGCs and Tor proviruses may integrate germline DNA after decapsidation. Right: PGCs become germ cells that proliferate in the adult gonad. New Tor copies resulting from earlier infection are now present in the germline genome (white boxes). Factors ensuring stringent silencing of TEs remain to be fully characterized in Oikopleura.
We found that Tor represents a unique case among LTR retrotransposons and retroviruses, in using an original mechanism for env transcription and exhibiting high levels of embryonic, somatic, tissue-specific expression. This very characteristic expression of Tor in embryos complements other observations of somatic expression of TEs during animal development (54–56). The anatomical simplicity of Oikopleura embryos should be an asset for further studies of soma-germline interactions in the proliferation of TEs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Jean-Marie Bouquet and the Oikopleura facility for excellent assistance in injection experiments and animal breeding, Dr Lisbeth Charlotte Olsen for providing the vas4 probe construct, Dr Gemma Danks for assistance in analysing the genome tiling array data. We also thank Dr Christophe Terzian, Université Claude Bernard in Lyon, for valuable discussions.
FUNDING
This work was supported by the University of Bergen and the Research Council of Norway [234817 to the Sars International Centre for Marine Molecular Biology]. Funding for open access charge: University of Bergen.
Conflict of interest statement. None declared.
REFERENCES
- 1.Eickbush T.H., Furano A.V. Fruit flies and humans respond differently to retrotransposons. Curr. Opin. Genet. Dev. 2002;12:669–674. doi: 10.1016/s0959-437x(02)00359-3. [DOI] [PubMed] [Google Scholar]
- 2.Redelsperger F., Cornelis G., Vernochet C., Tennant B.C., Catzeflis F., Mulot B., Heidmann O., Heidmann T., Dupressoir A. Capture of syncytin-Mar1, a fusogenic endogenous retroviral envelope gene involved in placentation in the rodentia squirrel-related clade. J. Virol. 2014;88:7915–7928. doi: 10.1128/JVI.00141-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delsuc F., Brinkmann H., Chourrout D., Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- 4.Robison B.H., Reisenbichler K.R., Sherlock R.E. Giant larvacean houses: rapid carbon transport to the deep sea floor. Science. 2005;308:1609–1611. doi: 10.1126/science.1109104. [DOI] [PubMed] [Google Scholar]
- 5.Bouquet J.M., Spriet E., Troedsson C., Ottera H., Chourrout D., Thompson E.M. Culture optimization for the emergent zooplanktonic model organism Oikopleura dioica. J. Plankton Res. 2009;31:359–370. doi: 10.1093/plankt/fbn132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sagane Y., Hosp J., Zech K., Thompson E.M. Cytoskeleton-mediated templating of complex cellulose-scaffolded extracellular structure and its association with oikosins in the urochordate Oikopleura. Cell. Mol. Life Sci. 2011;68:1611–1622. doi: 10.1007/s00018-010-0547-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denoeud F., Henriet S., Mungpakdee S., Aury J.M., Da Silva C., Brinkmann H., Mikhaleva J., Olsen L.C., Jubin C., Canestro C., et al. Plasticity of animal genome architecture unmasked by rapid evolution of a pelagic tunicate. Science. 2010;330:1381–1385. doi: 10.1126/science.1194167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Volff J.N., Lehrach H., Reinhardt R., Chourrout D. Retroelement dynamics and a novel type of chordate retrovirus-like element in the miniature genome of the tunicate Oikopleura dioica. Mol. Biol. Evol. 2004;21:2022–2033. doi: 10.1093/molbev/msh207. [DOI] [PubMed] [Google Scholar]
- 9.Malik H.S., Henikoff S., Eickbush T.H. Poised for contagion: evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res. 2000;10:1307–1318. doi: 10.1101/gr.145000. [DOI] [PubMed] [Google Scholar]
- 10.White J.M., Delos S.E., Brecher M., Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen C.H., Steiner L.A. Genome structure and thymic expression of an endogenous retrovirus in zebrafish. J. Virol. 2004;78:899–911. doi: 10.1128/JVI.78.2.899-911.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pelisson A., Song S.U., Prud'homme N., Smith P.A., Bucheton A., Corces V.G. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J. 1994;13:4401–4411. doi: 10.1002/j.1460-2075.1994.tb06760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tazi J., Bakkour N., Marchand V., Ayadi L., Aboufirassi A., Branlant C. Alternative splicing: regulation of HIV-1 multiplication as a target for therapeutic action. FEBS J. 2010;277:867–876. doi: 10.1111/j.1742-4658.2009.07522.x. [DOI] [PubMed] [Google Scholar]
- 14.Vicient C.M., Kalendar R., Schulman A.H. Envelope-class retrovirus-like elements are widespread, transcribed and spliced, and insertionally polymorphic in plants. Genome Res. 2001;11:2041–2049. doi: 10.1101/gr.193301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brasset E., Taddei A.R., Arnaud F., Faye B., Fausto A.M., Mazzini M., Giorgi F., Vaury C. Viral particles of the endogenous retrovirus ZAM from Drosophila melanogaster use a pre-existing endosome/exosome pathway for transfer to the oocyte. Retrovirology. 2006;3:e25. doi: 10.1186/1742-4690-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dupressoir A., Heidmann T. Germ line-specific expression of intracisternal A-particle retrotransposons in transgenic mice. Mol. Cell. Biol. 1996;16:4495–4503. doi: 10.1128/mcb.16.8.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ketting R.F. The many faces of RNAi. Dev. Cell. 2011;20:148–161. doi: 10.1016/j.devcel.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 18.Dinkel H., Van Roey K., Michael S., Davey N.E., Weatheritt R.J., Born D., Speck T., Kruger D., Grebnev G., Kuban M., et al. The eukaryotic linear motif resource ELM: 10 years and counting. Nucleic Acids Res. 2014;42:D259–D266. doi: 10.1093/nar/gkt1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 21.Sagane Y., Zech K., Bouquet J.M., Schmid M., Bal U., Thompson E.M. Functional specialization of cellulose synthase genes of prokaryotic origin in chordate larvaceans. Development. 2010;137:1483–1492. doi: 10.1242/dev.044503. [DOI] [PubMed] [Google Scholar]
- 22.Seo H.C., Edvardsen R.B., Maeland A.D., Bjordal M., Jensen M.F., Hansen A., Flaat M., Weissenbach J., Lehrach H., Wincker P., et al. Hox cluster disintegration with persistent anteroposterior order of expression in Oikopleura dioica. Nature. 2004;431:67–71. doi: 10.1038/nature02709. [DOI] [PubMed] [Google Scholar]
- 23.Danks G., Campsteijn C., Parida M., Butcher S., Doddapaneni H., Fu B., Petrin R., Metpally R., Lenhard B., Wincker P., et al. OikoBase: a genomics and developmental transcriptomics resource for the urochordate Oikopleura dioica. Nucleic Acids Res. 2013;41:D845–D853. doi: 10.1093/nar/gks1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Misseri Y., Labesse G., Bucheton A., Terzian C. Comparative sequence analysis and predictions for the envelope glycoproteins of insect endogenous retroviruses. Trends Microbiol. 2003;11:253–256. doi: 10.1016/s0966-842x(03)00119-7. [DOI] [PubMed] [Google Scholar]
- 25.Zimmer G., Trotz I., Herrler G. N-glycans of F protein differentially affect fusion activity of human respiratory syncytial virus. J. Virol. 2001;75:4744–4751. doi: 10.1128/JVI.75.10.4744-4751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Telesnitsky A., Goff S.P. In: Retroviruses. Coffin JM, Hughes SH, Varmus HE, editors. NY: Cold Spring Harbor; 1997. [PubMed] [Google Scholar]
- 27.Huang C.R., Burns K.H., Boeke J.D. Active transposition in genomes. Annu. Rev. Genet. 46:651–675. doi: 10.1146/annurev-genet-110711-155616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ray D.A., Batzer M.A. Reading TE leaves: new approaches to the identification of transposable element insertions. Genome Res. 2011;21:813–820. doi: 10.1101/gr.110528.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stach T., Winter J., Bouquet J.M., Chourrout D., Schnabel R. Embryology of a planktonic tunicate reveals traces of sessility. Proc. Natl. Acad. Sci. U.S.A. 2008;105:7229–7234. doi: 10.1073/pnas.0710196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith Z.D., Chan M.M., Mikkelsen T.S., Gu H., Gnirke A., Regev A., Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Extavour C.G., Akam M. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development. 2003;130:5869–5884. doi: 10.1242/dev.00804. [DOI] [PubMed] [Google Scholar]
- 32.Kalmykova A.I., Klenov M.S., Gvozdev V.A. Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Res. 2005;33:2052–2059. doi: 10.1093/nar/gki323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuramochi-Miyagawa S., Watanabe T., Gotoh K., Takamatsu K., Chuma S., Kojima-Kita K., Shiromoto Y., Asada N., Toyoda A., Fujiyama A., et al. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev. 2010;24:887–892. doi: 10.1101/gad.1902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ganot P., Bouquet J.M., Thompson E.M. Comparative organization of follicle, accessory cells and spawning anlagen in dynamic semelparous clutch manipulators, the urochordate Oikopleuridae. Biol. Cell. 2006;98:389–401. doi: 10.1042/BC20060005. [DOI] [PubMed] [Google Scholar]
- 35.Schlesinger S., Lee A.H., Wang G.Z., Green L., Goff S.P. Proviral silencing in embryonic cells is regulated by Yin Yang 1. Cell Rep. 2013;4:50–58. doi: 10.1016/j.celrep.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blass E., Bell M., Boissinot S. Accumulation and rapid decay of non-LTR retrotransposons in the genome of the three-spine stickleback. Genome Biol. Evol. 2012;4:687–702. doi: 10.1093/gbe/evs044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duvernell D.D., Pryor S.R., Adams S.M. Teleost fish genomes contain a diverse array of L1 retrotransposon lineages that exhibit a low copy number and high rate of turnover. J. Mol. Evol. 2004;59:298–308. doi: 10.1007/s00239-004-2625-8. [DOI] [PubMed] [Google Scholar]
- 38.Flot J.F., Hespeels B., Li X., Noel B., Arkhipova I., Danchin E.G., Hejnol A., Henrissat B., Koszul R., Aury J.M., et al. Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. Nature. 2013;500:453–457. doi: 10.1038/nature12326. [DOI] [PubMed] [Google Scholar]
- 39.Daugherty M.D., Malik H.S. Rules of engagement: molecular insights from host-virus arms races. Annu. Rev. Genet. 2012;46:677–700. doi: 10.1146/annurev-genet-110711-155522. [DOI] [PubMed] [Google Scholar]
- 40.Campbell M., Eng C., Luciw P.A. The simian foamy virus type 1 transcriptional transactivator (Tas) binds and activates an enhancer element in the gag gene. J. Virol. 1996;70:6847–6855. doi: 10.1128/jvi.70.10.6847-6855.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reuss F.U., Coffin J.M. Mouse mammary tumor virus superantigen expression in B cells is regulated by a central enhancer within the pol gene. J. Virol. 1998;72:6073–6082. doi: 10.1128/jvi.72.7.6073-6082.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pongoski J., Asai K., Cochrane A. Positive and negative modulation of human immunodeficiency virus type 1 Rev function by cis and trans regulators of viral RNA splicing. J. Virol. 2002;76:5108–5120. doi: 10.1128/JVI.76.10.5108-5120.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dumesic P.A., Madhani H.D. Recognizing the enemy within: licensing RNA-guided genome defense. Trends Biochem. sci. 2013;39:25–34. doi: 10.1016/j.tibs.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dumesic P.A., Natarajan P., Chen C., Drinnenberg I.A., Schiller B.J., Thompson J., Moresco J.J., Yates J.R. 3rd, Bartel D.P., Madhani H.D. Stalled spliceosomes are a signal for RNAi-mediated genome defense. Cell. 2013;152:957–968. doi: 10.1016/j.cell.2013.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schiff R., Itin A., Keshet E. Transcriptional activation of mouse retrotransposons in vivo: specific expression in steroidogenic cells in response to trophic hormones. Genes Dev. 1991;5:521–532. doi: 10.1101/gad.5.4.521. [DOI] [PubMed] [Google Scholar]
- 46.Bronner G., Taubert H., Jackle H. Mesoderm-specific B104 expression in the Drosophila embryo is mediated by internal cis-acting elements of the transposon. Chromosoma. 1995;103:669–675. doi: 10.1007/BF00344227. [DOI] [PubMed] [Google Scholar]
- 47.Brookman J.J., Toosy A.T., Shashidhara L.S., White R.A. The 412 retrotransposon and the development of gonadal mesoderm in Drosophila. Development. 1992;116:1185–1192. doi: 10.1242/dev.116.4.1185. [DOI] [PubMed] [Google Scholar]
- 48.Dion-Cote A.M., Renaut S., Normandeau E., Bernatchez L. RNA-seq reveals transcriptomic shock involving transposable elements reactivation in hybrids of young lake whitefish species. Mol. Biol. Evol. 2014;31:1188–1199. doi: 10.1093/molbev/msu069. [DOI] [PubMed] [Google Scholar]
- 49.Xiol J., Spinelli P., Laussmann M.A., Homolka D., Yang Z., Cora E., Coute Y., Conn S., Kadlec J., Sachidanandam R., et al. RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell. 2014;157:1698–1711. doi: 10.1016/j.cell.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 50.Yajima M., Gustafson E.A., Song J.L., Wessel G.M. Piwi regulates Vasa accumulation during embryogenesis in the sea urchin. Dev. Dyn. 2014;243:451–458. doi: 10.1002/dvdy.24096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shigenobu S., Kitadate Y., Noda C., Kobayashi S. Molecular characterization of embryonic gonads by gene expression profiling in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 2006;103:13728–13733. doi: 10.1073/pnas.0603767103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siomi M.C., Sato K., Pezic D., Aravin A.A. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 53.Teysset L., Burns J.C., Shike H., Sullivan B.L., Bucheton A., Terzian C. A Moloney murine leukemia virus-based retroviral vector pseudotyped by the insect retroviral gypsy envelope can infect Drosophila cells. J. Virol. 1998;72:853–856. doi: 10.1128/jvi.72.1.853-856.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baillie J.K., Barnett M.W., Upton K.R., Gerhardt D.J., Richmond T.A., De Sapio F., Brennan P.M., Rizzu P., Smith S., Fell M., et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mey A., Acloque H., Lerat E., Gounel S., Tribollet V., Blanc S., Curton D., Birot A.M., Nieto M.A., Samarut J. The endogenous retrovirus ENS-1 provides active binding sites for transcription factors in embryonic stem cells that specify extra embryonic tissue. Retrovirology. 2012;9:e21. doi: 10.1186/1742-4690-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perrat P.N., DasGupta S., Wang J., Theurkauf W., Weng Z., Rosbash M., Waddell S. Transposition-driven genomic heterogeneity in the Drosophila brain. Science. 2013;340:91–95. doi: 10.1126/science.1231965. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


