Figure 4.
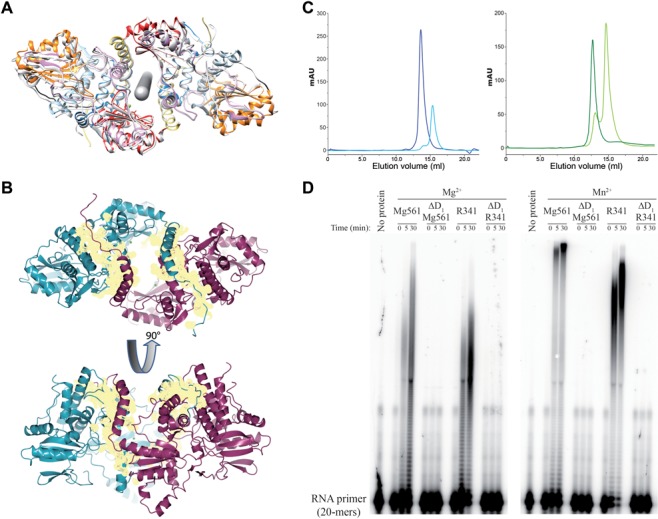
Molecular determinant of mimiviruses PAP dimerization and processivity. (A) Mg561 dimer symmetry. Rotational symmetry axis is shown in gray. Figure 1 color code is applied to one monomer and the second monomer is in light gray. (B) Buried surface area of the Mg561 dimer. The dimer interface buries a surface area (yellow) of about 3200 Å2, which corresponds to 11.5% of each monomer's solvent accessible surface area (about 27 500 Å2). The first monomer is in cyan, the second one is in magenta. Cation locations are marked as green balls. (C) Gel filtration analysis of ΔD1 H6c-Mg561 and ΔD1 H6c-R341 mutants. Overlay of UV traces of the full-length H6c-Mg561 (dark blue curve) and ΔD1 H6c-Mg561 (light blue curve) proteins (Left panel) and of the full-length H6c-R341 (dark green curve) and ΔD1 H6c-R341 (light green curve) proteins (Right panel). Full-length H6c-Mg561 and H6c-R341 eluted as a single peak corresponding to a dimeric state: apparent molecular masses were 116.8 and 176.7 kDa, respectively, versus the calculated ones, 125.7 and 136.8 kDa. At least 90% of the ΔD1 H6c-Mg561 and ΔD1 H6c-R341 protein eluted as monomers: apparent molecular masses were 57.8 and 71.1 kDa versus the calculated 57.7 and 63.4 kDa. GE Healthcare markers were used for calibration. (D) PolyA polymerase assay of H6c-Mg561, ΔD1 H6c-Mg561, H6c-R341 and ΔD1 H6c-R341 on a 20-mers RNA primer in the presence of Mg2+ or Mn2+ as catalytic ions. Time course is in min.
