Figure 1.
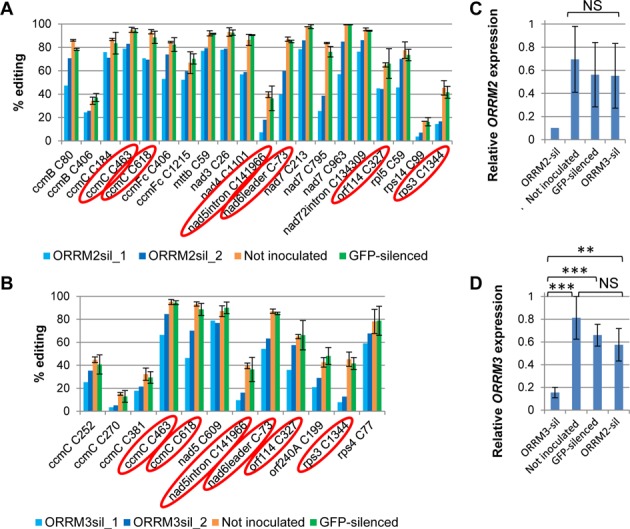
Transient silencing of ORRM2 and ORRM3 in Arabidopsis results in mitochondrial editing defects. Editing extent was measured by STS-PCRseq. ORRM2-sil, plants inoculated with Agrobacteria harboring a GFP and ORRM2 co-silencing construct. ORRM3-sil, plants inoculated with Agrobacteria harboring a GFP and ORRM3 co-silencing construct. Not inoculated, plants not inoculated with Agrobacteria. GFP-silenced, plants inoculated with Agrobacteria harboring a GFP silencing construct. Two replicates were used in each silencing experiment. Editing sites that experienced a decrease in editing extent ≥10% between the uninoculated controls and one of the silenced plants are shown. (A) Mitochondrial sites showing a reduction of editing extent in ORRM2-silenced plants. (B) Mitochondrial sites exhibiting a reduction of editing extent in ORRM3-silenced plants. Circledare the sites showing reduced editing extent in both silencing experiments. (C and D) Expression of silenced genes is measured by qRT-PCR. (C) Relative expression level of ORRM2 is reduced in ORRM2-silenced plants. (D) Relative expression level of ORRM3 is reduced in ORRM3-silenced plants. Significance ** (P < 0.01), *** (P < 0.001), NS (P ≥ 0.05).
