Abstract
The Merkel cell polyomavirus (MCPyV), discovered in 2008, drives development of most Merkel cell carcinomas (MCCs) through several canonical mechanisms. A glaring gap in our knowledge remains the basis by which MCPyV, among all 12 human polyomaviruses, is the only one that causes cancer in humans. Moreover, initial attempts by numerous groups have failed to reproduce MCC in mice using oncoproteins from this polyomavirus. Verhaegen at al. report MCPyV small T antigen-expressing transgenic mice that now provide insight into in vivo transformation mechanisms.
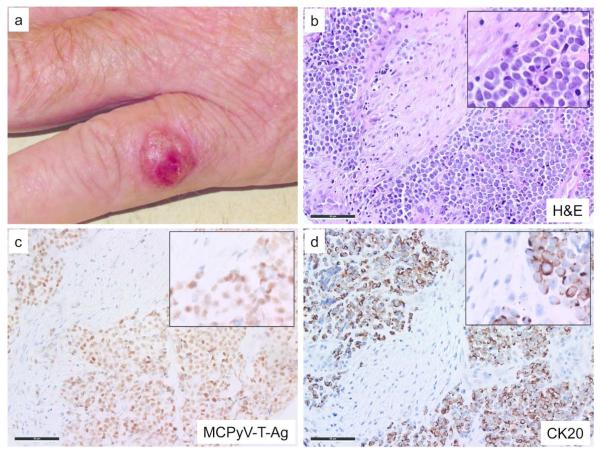
Merkel cell carcinoma is a rare and aggressive human neuroendocrine skin cancer with a disease-associated mortality of approximately 50% (Lemos et al., 2010). Although reported incidences are increasing rapidly, it remains an orphan disease, with approximately 1600 new cases per year in the United States. Accordingly, until recently, little was known about its pathogenesis and few clinical trials were available. MCC is associated with clonal integration of the Merkel cell polyomavirus in about 80% of cases (Feng et al., 2008), while a subset of cases appear to be truly independent of this virus. MCC typically arises on the UV-exposed skin of Caucasian individuals, and it often has a clinically benign appearance that may resemble a painless nodular cyst. Figure 1 shows representative clinical and microscopic features of a Merkel cell carcinoma.
Figure 1. Clinical and microscopic images of Merkel cell carcinoma.
a) Representative MCC on the left hand of a 70-year-old man. Microscopic images with magnified insets of a primary MCC tumor with b) Hematoxylin and eosin stain showing salt and pepper chromatin pattern, frequent mitotic figures and nuclear molding characteristic of MCC; c) MCPyV Large T-Ag immunohistochemistry (CM2B4 antibody) shows viral protein expression in tumor cells but not adjacent stroma; d) Cytokeratin 20 (CK20) immunohistochemistry demonstrates characteristic dot-like peri-nuclear staining. Scale bar, 50 μM
Development of MCC involves several molecular steps, including upregulation of cell cycle progression proteins such as cyclin-E, inactivation of tumor suppressor proteins such as retinoblastoma (Rb) (Sihto et al., 2011) as well as immune evasion (Afanasiev et al., 2013a; Paulson et al., 2014). Great insight into the biology of this cancer has been obtained through characterization of human tumor material, including MCC tumor cell lines. However, despite extensive effort, no animal model has been developed to determine how individual MCPyV oncoproteins interact with host cell pathways in vivo.
In this issue of the Journal of Investigative Dermatology, Verhaegen at al., demonstrate that expression of MCPyV small T-antigen (sT-Ag) results in dysregulation in epidermal differentiation in embryonic mice and in squamous cell carcinoma in situ when inducibly expressed in adult mice (Verhaegen et al., 2014). Furthermore, the authors demonstrate that the “LSD” (Large T-antigen Stabilization Domain) of the sT-Ag is necessary for transformation by this oncoprotein via the ubiquitin E3 ligase pathway.
Oncoproteins from the Merkel virus: what do we know?
A watershed finding regarding the etiology of MCC was the discovery in 2008 of the Merkel cell polyomavirus by the Chang-Moore group (Feng et al., 2008). They demonstrated that in most MCCs the viral DNA was clonally integrated into the host cell DNA, indicating that each tumor arose from a single cell and that the virus likely played an important etiologic role.
We now know that exposure to MCPyV is nearly ubiquitous, with antibodies specific to the capsid proteins of this virus often arising early in childhood (Chen et al., 2010). In contrast, development of MCC is rare and typically delayed by six to seven decades following initial virus exposure. Indeed, the presence of MCPyV does not lead directly to any apparent human disease, and this virus is often present on normal skin. Our current understanding of how this virus leads to MCC involves a “perfect storm” of events that is typically catalyzed by UV radiation exposure, and it includes integration of viral DNA, expression of small T antigen (sT-Ag), truncation/expression of large T-antigen (tLT-Ag), and evasion of a destructive immune response. When the above criteria are met and MCC develops, it is several times more likely to be fatal than any of the other more common skin cancers, including melanoma.
While MCPyV is ubiquitous and detection of its DNA by PCR can thus be difficult to interpret, immunohistochemistry data demonstrate persistent expression of MCPyV oncoproteins in MCC tumors, and the nature of viral DNA integration indicates that this virus is not merely a bystander. Consistent with the observation that sT-Ag and tLT-Ag are expressed persistently in human MCC tumor material, they appear to have separate and specific roles that are necessary for the ongoing growth of MCCs in vitro. Specifically, knocking down the sT-Ag and tLT-Ag oncoproteins results in growth arrest of MCPyV-driven MCC cells, indicating that ongoing expression of these oncoproteins is required (Houben et al., 2010). However, there has been debate in the field. For instance, the importance of sT-Ag in MCPyV mediated transformation has been controversial, as one research group reported that knockdown of sT-Ag leads to growth arrest of MCC cells (Shuda et al., 2014; Shuda et al., 2011) while another group found that sT-Ag is dispensable for growth (Angermeyer et al., 2013). Additionally, MCPyV sT-Ag has the ability to transform cells independent of LT-Ag, which is not the case for the SV40 polyomavirus (Shuda et al., 2011).
In vitro experiments to understand the molecular mechanisms of carcinogenesis have elucidated several transforming pathways (shared among polyomaviruses), but it remains unclear why MCPyV is the only polyomavirus known to cause cancer in humans. A fascinating and critical finding is that the LT-Ag is invariably truncated in MCC but that the precise location and nature of the truncation varies from tumor to tumor (Shuda et al., 2008). Importantly however, the truncation event in LT-Ag is always downstream of the Rb domain (Fig. 2; this retains Rb inactivating activity and promotes cell cycle progression), and upstream of the helicase domain (which renders the viral origin irrelevant, thus eliminating virion production which would otherwise be fatal for the cell (Shuda et al., 2008)).
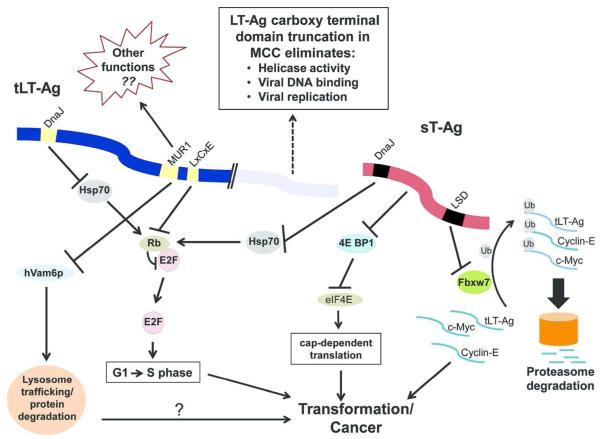
Figure 2. MCPyV T-Antigen oncoprotein functional interactions with cellular pathways.
The MCPyV small T-antigen (sT-Ag) and truncated Large T-antigen (tLT-Ag) are depicted together with their major known cellular targets. The DnaJ region is present in sT-Ag and LT-Ag amino terminal regions and interacts with heat shock protein 70 (Hsp70). Hsp70 binding by sT-Ag and LT-Ag indirectly inactivates the tumor suppressor protein Rb (Houben et al., 2014) thereby promoting progression from G1 to S phase. The MUR1 domain of truncated LT-Ag binds and inactivates hVam6p. This inactivation leads to disruption of lysosome trafficking, allowing accumulation of certain proteins in the host cell (Liu et al., 2011), possibly contributing to oncogenesis. The MUR1 domain may have other functions not yet characterized that could help explain why MCPyV is carcinogenic in humans. Truncated LT-Ag also binds and inactivates Rb through the LxCxE motif, preventing it from regulating the cell cycle (Houben et al., 2014) and promoting progression from G1 to S phase. sT-Ag prevents the turnover of hyperphosphorylated 4E-BP1, releasing activated eIF4E and increasing cap dependent translation which contributes to cell transformation (Shuda et al., 2011). The sT-Ag LSD domain binds Fbxw7, inhibits its ubiquitin ligase activity and thus promotes the stability of critical oncogenic proteins including LT-Ag, c-Myc and cyclin-E (Kwun et al., 2013).
Perturbing regulatory proteins and circumventing cell cycle progression checkpoints are necessary to drive tumorgenesis, and these are commonly mediated by viral or cellular oncoproteins. A striking example in MCC is Rb inactivation, which is clearly essential for MCC development (Sihto et al., 2011), and this is achieved through different mechanisms in MCCs that are MCPyV-positive (via T-antigen binding) compared with those that are MCPyV–negative (via mutation of Rb). A simplified version of the relevant cell transformation pathways that are affected by MCPyV oncoproteins is shown in Figure 2.
MCPyV has two regions that are not present in other polyomaviruses, MCPyV Unique Region 1 and 2 (MUR1 & MUR2). MUR2 is not expressed in MCC due to truncation of the LT-Ag, but MUR1 is retained in truncated LT-Ag and thus could mediate some of the unique biological properties of this virus. MUR1 contains a domain capable of binding hVam6p, a protein involved in lysosomal trafficking. When hVam6p is sequestered in HIV-1 infected cells, lysosomal protein degradation is reduced and viral progeny production is increased (Molle et al., 2009). Although inhibition of hVam6p can thus be important in the lifecycle of a virus, it is uncertain whether this protein plays a role in MCC (Houben et al., 2014).
To date, there have been no reports of transgenic mouse models in which MCPyV oncoproteins drive carcinogenesis successfully. In contrast, many other polyomaviruses readily cause cancer in rodents, helping to explain why this family of viruses was named “poly-oma” (many tumors).
At last, an MCPyV oncoprotein mouse model
For decades, the polyomavirus T-Ags (specifically those from SV40) have been powerful tools in deciphering the molecular mechanisms of tumorgenesis and mammalian cell biology. Indeed, the use of SV40 T-Ag to transform cells led to fundamental discoveries such as the roles of the Rb and p53 tumor suppressors (Pipas, 2009). However, the inability to generate a MCPyV-driven animal model is surprising and has hindered progress in the field.
Verhaegen et al., have now characterized transgenic mice that constitutively express wild type sT-Ag under the control of an epidermis-specific promoter (keratin-5). These mice expressed sT-Ag within the stratified squamous epithelia and developed striking epithelial dysplasia. The authors used amino acid substitution to disrupt several sT-Ag domains in order to probe their relative contributions. When the “LSD” (Large T-antigen Stabilization Domain) of sT-Ag was mutagenized, transgenic mice no longer developed epithelial dysplasia. The authors showed that this phenotype was likely due to dysregulation of protein degradation. Specifically, the LSD binds Fbxw7, an E3 ubiquitin ligase that forms a complex responsible for tagging proteins destined for the proteasome. Upon binding Fbxw7, sT-Ag inactivates this ubiquitin ligase by sequestering it in the cell nucleus, leading to accumulation of oncogenic proteins such as cyclin-E, c-Jun, mTOR, and truncated LT-Ag (Verhaegen et al., 2014). These in vivo findings agree with earlier studies that demonstrated LSD function to promote cellular transformation and to support survival of MCC cell lines (Kwun et al., 2013).
Verhaegen et al., note that they “analyzed pre-term transgenic embryos to circumvent a potentially severe phenotype incompatible with postnatal survival”, indicating that constitutive expression of the sT-Ag was lethal. The toxicity of this transgene underscores the difficulty of studying MCPyV sT-Ag-driven transformation. Accordingly, the authors characterized the inducible expression of sT-Ag in adult mice, and they found that this leads to a phenotype that strongly resembles squamous cell carcinoma (SCC) in situ. Indeed, “collision tumors” between MCC and squamous cell carcinoma (SCC) in situ (Bowen’s Disease) have been reported several times in the literature, suggesting that they may arise from a common neoplastic precursor lesion. Histological examination of such collision tumors often reveals pleomorphic keratinocytes, characteristic of SCC in situ, admixed with cells with an MCC phenotype (hyperchromatic nuclei and peri-nuclear CK20 staining) (Sirikanjanapong et al., 2010). These lesions frequently occur on sun-exposed skin implicating UV damage as the underlying etiology of disease, and interestingly, are almost invariably negative for MCPyV. The fact that the inducible MCPyV sT-Ag mouse model of Verhaegen et al. develops SCC-like lesions also suggests there are shared pathways between virus-and UV-driven carcinogenesis.
While this inducible mouse model does not fully recapitulate MCC, it demonstrates that the LSD domain is necessary for sT-Ag-driven transformation of cells in vivo and provides a valuable tool to the field for exploring biology and possible therapeutic approaches.
How might recent pathogenic insights help patients?
Small molecule inhibitors that selectively target viral proteins or their cellular targets could provide opportunities to disrupt virus-driven carcinogenesis and to “translate” these mechanistic findingsdirectly toward clinical benefit. The Chang-Moore group identified a potent inhibitor of MCC, YM155 (Arora et al., 2012). YM155 inhibits translation of survivin, an oncoprotein that is up-regulated in MCC regardless of MCPyV status, in addition to many other cancers. In a xenograft mouse model, YM155 strikingly slowed the growth of MCC tumors and induced non-apoptotic cell death in MCC cell lines. Accordingly, YM155 has been tested in several phase I/II clinical trials for treatment of various cancers but a clinical trial of YM155 in MCC has not yet come to fruition. The inducible sT-Ag mouse model developed by Verhaegen et al., could be a useful model to perform small-molecule discovery and toxicity studies to expand opportunities for therapeutic targets in MCC.
A characteristic of virus-driven cancers is the expression of non-self, viral proteins that should be readily detectable by the immune system. Numerous lines of evidence demonstrate that immune function is important for recognizing and eliminating MCC. Specifically, solid organ transplant recipients, HIV/AIDS patients, and those with hematologic malignancies, are at higher risk for developing MCC, and they have poorer outcomes. However, while over 90% of MCC patients have no known immune dysfunction, they fail to eliminate these tumors that persistently express MCPyV oncoproteins. How do these highly antigenic tumors evade immunological destruction? Indeed, several immune evasion mechanisms appear to be active in MCC, and in some cases are reversible. Specifically, over 80% of MCC tumors down-regulate the expression of MHC class I (Paulson et al., 2014), thereby suppressing immune recognition of MCPyV-derived peptides by CD8 T cells. Additionally, vascular E-selectin expression is reduced in many MCC tumors, effectively diminishing the ability of lymphocytes to migrate into the tumor microenvironment (Afanasiev et al., 2013a). When the cellular immune response is not successfully subverted by MCC (as assessed by CD8+ lymphocytes found within the tumor), 100% disease-specific survival ensues, even in patients presenting with advanced nodal or distant metastatic disease (Paulson et al., 2011).
According to Clinicaltrials.gov, there are currently 17 active clinical trials that are specifically designed to include MCC patients. Seven of these trials involve immunotherapies that aim to augment anti-tumor immune responses. Indeed, MCPyV-specific T cells have been shown to express elevated levels of multiple markers of exhaustion such as PD-1 and TIM-3 (Afanasiev et al., 2013b). Trials are active that target the CD8 T cell response to reverse T cell exhaustion via antibodies to PD-1 or PD-L1. The focus on immune-based trials in MCC reflects the striking advances in the field of cancer immunology, which may be especially relevant for a virus-driven malignancy.
In summary, the past few years have provided remarkable insights into how MCPyV drives this cancer and how the immune system should typically control it. These diverse insights promise to provide us with a more comprehensive toolbox with which to treat MCC patients who currently have limited therapeutic options.
Pullquote.
We still do not know why MCPyV, among all 12 human polyomaviruses, is the only one that causes cancer.
Acknowledgements
Funding sources: T32-AR056969, K24-CA139052, R01-CA176841, R01-CA162522, MCC patient gift fund. We thank Natalie Miller and Natalie Vandeven for helpful discussions and Chris Lewis and Teresa Fu for assistance with figure preparation.
References
- Afanasiev OK, Nagase K, Simonson WT, et al. Vascular E-selectin expression correlates with CD8 lymphocyte infiltration and improved outcome in Merkel cell carcinoma. Journal of Investigative Dermatology. 2013a;133:2065–73. doi: 10.1038/jid.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afanasiev OK, Yelistratova L, Miller N, et al. Merkel polyomavirus-specific T cells fluctuate with merkel cell carcinoma burden and express therapeutically targetable PD-1 and Tim-3 exhaustion markers. Clinical Cancer Research. 2013b;19:5351–60. doi: 10.1158/1078-0432.CCR-13-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angermeyer S, Hesbacher S, Becker JC, et al. Merkel cell polyomavirus-positive Merkel cell carcinoma cells do not require expression of the viral small T antigen. Journal of Investigative Dermatology. 2013;133:2059–64. doi: 10.1038/jid.2013.82. [DOI] [PubMed] [Google Scholar]
- Arora R, Shuda M, Guastafierro A, et al. Survivin Is a Therapeutic Target in Merkel Cell Carcinoma. Science translational medicine. 2012;4:133ra56. doi: 10.1126/scitranslmed.3003713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Hedman L, Mattila PS, et al. Serological evidence of Merkel cell polyomavirus primary infection in childhood. Journal of Clinical Virology. 2010;50:125–9. doi: 10.1016/j.jcv.2010.10.015. [DOI] [PubMed] [Google Scholar]
- Feng H, Shuda M, Chang Y, et al. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science (New York, NY) 2008;319:1096–100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben R, Angermeyer S, Haferkamp S, et al. Characterization of functional domains in the Merkel cell polyoma virus Large T antigen. International Journal of Cancer. 2014 doi: 10.1002/ijc.29200. Advanced online publication, 10 September 2014. (doi: 10.1002/ijc.29200) [DOI] [PubMed] [Google Scholar]
- Houben R, Shuda M, Weinkam R, et al. Merkel Cell Polyomavirus-Infected Merkel Cell Carcinoma Cells Require Expression of Viral T Antigens. Journal of virology. 2010;84:7064–72. doi: 10.1128/JVI.02400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwun HJ, Shuda M, Feng H, et al. Merkel cell polyomavirus small T antigen controls viral replication and oncoprotein expression by targeting the cellular ubiquitin ligase SCFbw7. Cell host & microbe. 2013;14:125–35. doi: 10.1016/j.chom.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos BD, Storer BE, Iyer JG, et al. Pathologic nodal evaluation improves prognostic accuracy in Merkel cell carcinoma: analysis of 5823 cases as the basis of the first consensus staging system. Journal of the American Academy of Dermatology. 2010;63:751–61. doi: 10.1016/j.jaad.2010.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Hein J, Richardson SCW, et al. Merkel Cell Polyomavirus Large T Antigen Disrupts Lysosome Clustering by Translocating Human Vam6p from the Cytoplasm to the Nucleus. Journal of Biological Chemistry. 2011;286:17079–90. doi: 10.1074/jbc.M110.192856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molle D, Segura-Morales C, Camus G, et al. Endosomal trafficking of HIV-1 gag and genomic RNAs regulates viral egress. Journal of Biological Chemistry. 2009;284:19727–43. doi: 10.1074/jbc.M109.019844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson KG, Iyer JG, Tegeder AR, et al. Transcriptome-wide studies of Merkel cell carcinoma and validation of intratumoral CD8+ lymphocyte invasion as an independent predictor of survival. Journal Clinical Oncology. 2011;29:1539–46. doi: 10.1200/JCO.2010.30.6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson KG, Tegeder A, Willmes C, et al. Downregulation of MHC-I expression is prevalent but reversible in Merkel cell carcinoma. Cancer immunology research. 2014;2:1071–9. doi: 10.1158/2326-6066.CIR-14-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipas JM. SV40: Cell transformation and tumorigenesis. Virology. 2009;384:294–303. doi: 10.1016/j.virol.2008.11.024. [DOI] [PubMed] [Google Scholar]
- Shuda M, Chang Y, Moore PS. Merkel cell polyomavirus positive Merkel cell carcinoma requires viral small T antigen for cell proliferation. Journal of Investigative Dermatology. 2014;134:1479–81. doi: 10.1038/jid.2013.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuda M, Feng B, Kwun HJ, et al. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. PNAS. 2008;105:16272–7. doi: 10.1073/pnas.0806526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuda M, Kwun HJ, Feng H, et al. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E BP1 translation regulator. Journal of Clinical Investigation. 2011;121:3623–34. doi: 10.1172/JCI46323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sihto H, Kukko H, Koljonen V, et al. Merkel cell polyomavirus infection, large T antigen, retinoblastoma protein and outcome in Merkel cell caarcinoma. Clinical Cancer Research. 2011;17:4806–13. doi: 10.1158/1078-0432.CCR-10-3363. [DOI] [PubMed] [Google Scholar]
- Sirikanjanapong S, Melamed J, Patel RR. Intraepidermal and dermal Merkel cell carcinoma with squamous cell carcinoma in situ: a case report with review of literature. Journal of cutaneous pathology. 2010;37:881–5. doi: 10.1111/j.1600-0560.2009.01407.x. [DOI] [PubMed] [Google Scholar]
- Verhaegen ME, Mangelberger D, Harms PW, et al. Merkel cell polyomavirus small T antigen is oncogenic in transgenic mice. Journal of Investigative Dermatology. 2014 doi: 10.1038/jid.2014.446. [DOI] [PMC free article] [PubMed] [Google Scholar]