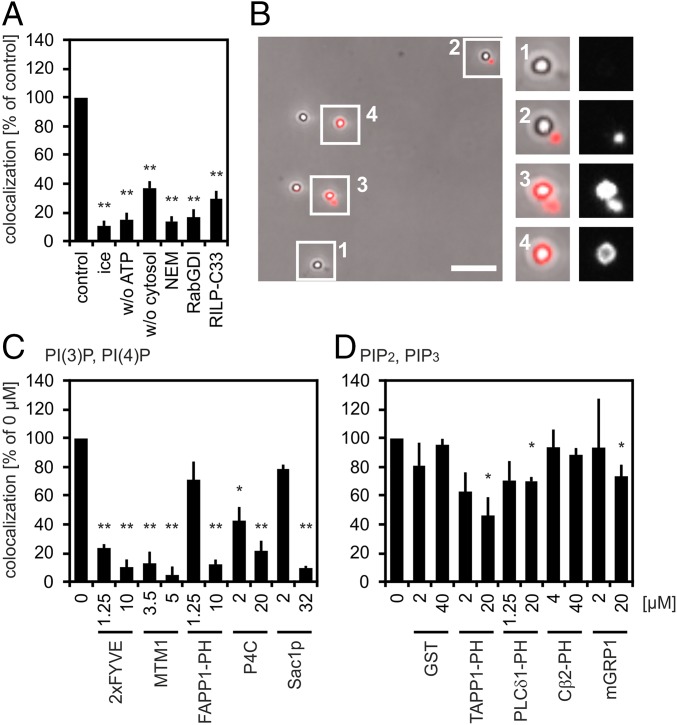
Fig. 1.
PLF requires PI(4)P and PI(3)P. (A) Phagosomes containing 1-µm latex beads (LBPs) and lysosomes containing BSA-rho-bio were mixed under fusion-promoting conditions. LBPs were isolated from reaction mixtures, spun onto coverslips, and assayed for colocalization with the lysosomal fluor. Colocalization under standard conditions (control) was set as 100% (4–10% absolute colocalization). Reactions were incubated for 60 min on ice or at 37 °C in the absence of ATP or cytosol or in the presence of 4 mM N-ethylmaleimide (NEM), 10 µM RabGDI, or 6 µM Rab7-interacting lysosomal protein-C33 (RILP-C33). (B) Micrograph showing LBPs from a cell-free fusion reaction prepared for fluorescence microscopy. Fusion of LBPs with lysosomes results in colocalization between LBPs and lysosomal BSA-rho-bio. Enlargements show LBPs colocalizing (nos. 3 and 4) or not colocalizing (nos. 1 and 2) with the lysosomal fluor. (Scale bar, 5 µm.) Enlargements (2.5-fold) show LBPs colocalizing (3 and 4) or not colocalizing. (C and D) Recombinant proteins were added to standard fusion reactions as indicated. Colocalization in the absence of purified proteins (0 µM) was defined as 100% (4–20% of phagosomes containing the lysosomal tracer). Data are means ± SEM from at least three independent experiments. Proteins added to fusion reactions were: (C) PI(3)P binder 2xFYVE, PI(3)P phosphatase MTM1, PI(4)P binders FAPP1 PH domain and SidC P4C, or PI(4)P phosphatase Sac1p; (D) binders to poly-phosphorylated PIPs (PI(3,4)P2, TAPP1 PH domain; PI(4,5)P2, PLCδ1 PH domain; PI(3,5)P2, Centaurinß2 PH domain (Cβ2-PH); PI(3,4,5)P3, mGRP1) or GST. Final protein concentrations are indicated. *P < 0.05, **P < 0.01 compared with untreated controls (= control, = 0 µM).