Abstract
The malaria parasite Plasmodium falciparum exports a large number of proteins into the erythrocyte cytoplasm during the asexual intraerythrocytic stage of its life cycle. A subset of these proteins interacts with erythrocyte membrane skeletal proteins and grossly alters the structure and function of the membrane. Several of the exported proteins, such as PfEMP1, PfEMP3, RESA and KAHRP, interact with the preponderant erythrocyte skeleton protein, spectrin. Here we have searched for possible interaction of these four malaria proteins with another major erythrocyte skeleton protein, ankyrin R. We have shown that KAHRP, but none of the other three, binds to ankyrin R. We have mapped the binding site for ankyrin R to a 79-residue segment of the KAHRP sequence, and the reciprocal binding site for KAHRP in ankyrin R to a subdomain (D3) of the 89 kDa ankyrin R membrane-binding domain. Interaction of intact ankyrin R with KAHRP was inhibited by the free D3 subdomain. When, moreover, red cells loaded with the soluble D3 subdomain were infected with P. falciparum, KAHRP secreted by the intraerythrocytic parasite no longer migrated to the host cell membrane, but remained diffusely distributed throughout the cytosol. Our findings suggest a potentially important role for interaction of KAHRP with red cell membrane skeleton in promoting the adhesion of malaria-infected red cells to endothelial surfaces, a central element in the pathophysiology of malaria.
Keywords: Red cell, Malaria, Cytoskeletal proteins, Ankyrin R, KAHRP
1. Introduction
Plasmodium falciparum causes the most severe form of human malaria and is responsible for nearly all malaria-induced mortality. During the asexual intraerythrocytic life cycle of P. falciparum, the parasite exports a large number of proteins into the red cell cytoplasm, some of which bind to the host membrane skeleton proteins [1–3]. The resulting interactions induce extensive structural and functional changes in the infected erythrocytes, notably increased adhesiveness to endothelial surfaces, decreased membrane deformability and altered cell morphology and elevated permeability to a wide variety of ions [4–6]. To determine the mechanisms by which the parasite proteins exert their effects on the host cell, we and others have sought to define the nature of their interactions with the components of the host cell membrane. To date, five P. falciparum proteins have been reasonably well characterized, namely erythrocyte membrane-binding protein 1(PfEMP1), the erythrocyte membrane-binding protein 3 (PfEMP3), the ring parasite-infected erythrocyte surface antigen (RESA), the knob-associated histidine-rich protein (KAHRP), and the mature parasite-infected erythrocyte surface antigen (MESA).
The erythrocyte membrane is a composite structure in which the lipid bilayer with its integral proteins is linked to a two-dimensional membrane skeleton network. The major skeleton proteins are α-spectrin, β-spectrin, actin, protein 4.1R, ankyrin R, protein 4.2, p55, adducin, dematin, tropomyosin, and tropomodulin [7–9]. The α- and β-spectrins in the form of predominantly α2β2 tetramers are the principal components of the membrane skeletal network [10,11], which is attached to the lipid bilayer through two pathways, both involving integral and peripheral protein constituents. One involves ankyrin R, which interacts with the cytoplasmic domain of the preponderant transmembrane protein, band 3, forming the “ankyrin R-complex”, which is attached to the spectrin tetramers at a point near the point of apposition of the two αβ dimers [12]. The other involves protein 4.1R which associates with the transmembrane glycophorin C to form the “4.1R-complex” at the spectrin–actin junctional complex at the network nodes [13].
Of the five relatively well-studied exported malarial proteins RESA, KAHRP, PfEMP1, PfEMP3 and MESA, all but MESA have been shown to interact with spectrin. Each of these four proteins has its own separate binding site in spectrin, and the consequences of their interactions are accordingly distinguishable. While RESA binds to β-spectrin repeat 16, close to the labile dimer–dimer self-association site, PfEMP3 binds to the EF hands of α-spectrin at the distal ends of the tetramers. Consequently, the RESA–spectrin interaction stabilizes the spectrin tetramer against dissociation into its constituent dimers, thereby increasing shear resistance of the membrane stability [14]. In contrast, PfEMP3 destabilizes the membrane by dissociating the spectrin–actin–4.1 ternary complex [15]. KAHRP binds to repeat 4 of α-spectrin. Although KAHRP–α-spectrin association has no effect on the membrane mechanical properties, interaction of KAHRP with spectrin is required for the proper assembly of KAHRP into the knob complex found at the erythrocyte membrane [16].
In contrast to the well-characterized interactions of malaria proteins with spectrin, their relation to other major erythrocyte skeleton proteins is less well understood. To date, MESA has been reported to bind exclusively protein 4.1R [17–19]. In vivo studies support the role of trafficking and skeletal-binding motifs in the interaction of MESA with the membrane skeleton of P. falciparum-infected red blood cells [20]. MESA–4.1R interaction appears to be important for intraerythrocytic growth of the parasite, since the invasion, growth and viability of MESA(+) parasites is reduced in 4.1R-deficient erythrocytes [21,22].
We have previously reported a putative association between KAHRP and ankyrin R based on immunoprecipitation of solubilized malaria-infected red blood cells [23]. In that study an antibody against KAHRP co-precipitated an 89 kDa protein fragment that was also recognized by an anti-ankyrin R antibody. Because of the nature of such studies an interaction can only be regarded as provisional. Also there was no study to examine the functional consequences of such an association. In the present study, we have searched for the interaction of malarial proteins with ankyrin R that bridges the membrane skeleton to the plasma membrane. We found that of the four malaria proteins we examined, only KAHRP bound to ankyrin R. We identified the ankyrin R-binding site in KAHRP and the reciprocal KAHRP-binding site in ankyrin R. The ankyrin R binding site is located within a 79 aa fragment of KAHRP. The fragment binds to the D3 region in the membrane-binding domain of ankyrin R. Furthermore, enclosure of the D3 poly-peptide in erythrocytes before infection by malarial parasites prevented the migration of KAHRP to the membrane. These findings imply that the interaction with ankyrin R is necessary for the attachment of KAHRP to the host cell membrane.
2. Material and methods
2.1. Materials
Type A fresh blood was obtained from healthy volunteers with informed consent. Serum from type A blood donor was obtained from the Interstate Blood Bank (Memphis, TN). Parasite clone 3D7 was obtained from MR4 (Manassas, VA). pGEX-4T-2 vector and glutathione-Sepharose 4B were purchased from Amersham Biosciences. pMAL-c2x vector, amylose resin and BL21(DE3) were from New England Bio-Lab. The reduced form of glutathione, isopropyl-β-D-galactopyranoside and Percoll were purchased from Sigma. Rabbit anti-ankyrin R, anti-KAHRP, anti-MBP and anti-GST antibodies were produced by our laboratory. HRP-conjugated anti-rabbit IgG was from Jackson Immuno Research Laboratory (West Grove, PA). FITC-conjugated donkey anti-rabbit IgG was from Invitrogen (Carlsbad, CA). The CM-5 sensor chip, amino coupling kit, and other reagents for SPR assay were purchased from BIAcore (Piscataway, NJ). All other chemicals were reagent grade products from standard sources.
2.2. Recombinant malaria and ankyrin R fragments
The recombinant GST-tagged KAHRP fragments (K1A, K1B, K1C, K1D, K2 and K3), MBP-tagged RESA fragments (F1, F2, F3, F4, F5, F6 and F7), GST-tagged PfEMP3 fragments (FIa, FIb, FII, FIII, FIV and FV), and GST-tagged cytoplasmic domain of PfEMP1 (VARC) have been previously described [14–16,24,25]. The three functional domains of ankyrin R (N-terminal 89 kDa membrane binding domain, internal 62 kDa spectrin-binding domain and C-terminal 55 kDa regulatory domain) and the subdomains of the membrane-binding domain (D1, D2, D3 and D4) were sub-cloned into pMAL-c2x vector using EcoRI and SalI upstream and downstream respectively. cDNA from human erythroblasts cultured from CD34+ cells was used as template for amplifying ankyrin fragments. The fidelity of all the constructs was verified by DNA sequencing.
2.3. Preparation of ankyrin R and recombinant proteins
Full length ankyrin R was purified from the red blood cells according to the method described by Tyler et al. [26]. To obtain recombinant proteins, cDNA encoding the desired polypeptide was transformed into the BL21 bacteria strain. The expression of recombinant proteins was induced by 0.1 mM isopropyl β-D-thiogalactopyranoside at 16 °C overnight. The GST-tagged polypeptides were purified using a glutathione-Sepharose 4B affinity column. The MBP-tagged poly-peptides were purified on an amylose resin affinity column. Proteins were dialyzed against PBS (10 mM phosphate, pH 7.4, 150 mM NaCl) for pull-down assays, against HBS-EP buffer (20 mM HEPES, pH7.4, 150 mM NaCl, 3 mM EDTA, and 0.05% surfactant P20) for surface plasmon resonance assay, or against hypotonic buffer (5 mM Tris, 5mMKCl, pH7.4) for resealing. Protein concentrations were determined by BCA kit (Thermo scientific, CA), using BSA as standard. All the proteins were clarified by ultracentrifugation at 230,000 g for 30 min at 4 °C before use.
2.4. Pull-down assays
To examine the binding of full-length ankyrin R to malaria proteins, recombinant GST-tagged KAHRP polypeptides, GST-tagged PfEMP3 polypeptides and the GST-tagged cytoplasmic domain of PfEMP1 were coupled to glutathione-Sepharose 4B beads in a total volume of 100 μl (at the concentration of 1 μM) at room temperature for 30 min. MBP-tagged RESA fragments were coupled to amylase beads. Beads were pelleted and washed, then ankyrin R, at a concentration 1 μM in a volume of 100 μl, was added to the beads. The mixture was incubated for 1 h at room temperature, pelleted, washed, and eluted with 10% SDS. The pellet was analyzed by SDS-PAGE. The binding of ankyrin R to KAHRP fragments was detected by Western blot analysis using an anti-ankyrin R antibody. To examine the binding between recombinant KAHRP fragments and ankyrin R fragments, MBP-tagged ankyrin R fragments were coupled to amylose beads. After washing, GST-tagged KAHRP K1D fragment was added. The binding was detected by Western blot using an anti-GST antibody. Equal coupling of each tagged protein to the beads was validated by SDS-PAGE followed by CBB staining.
2.5. Enzyme-linked immunosorbent assay (ELISA)
ELISA was used for detecting direct binding between ankyrin R and KAHRP and for the inhibition assay. To examine the binding of full length ankyrin R to GST-tagged KAHRP fragments, full length ankyrin R (0. 1 μM) was coated on the 96-well plate overnight at 4 °C. The plate was washed and blocked with 1% BSA in PBS, with 0.05% Tween 20, for 1 h at room temperature. GST-tagged K1A, K1B, K1C, K1D, K2 and K3 (each 0. 1 μM) was added and the reaction was continued for another 30 min. The binding of GST-tagged K1D was detected by an anti-GST antibody. The plate was read at 450 nm. For inhibition assay, full length ankyrin R (0. 1 μM) was coated on the 96-well plate overnight at 4 °C. The plate was washed and blocked with 1% BSA in PBS, with 0.05% Tween 20, for 1 h at room temperature. GST-tagged K1D (0.1 μM) pre-incubated with various concentrations (0 μM–8 μM) of MBP-D3 (MBP as control) was added and the reaction was continued for another 30 min. The binding of GST-tagged K1D was detected by an anti-GST antibody. The plate was read at 450 nm.
2.6. Surface plasmon resonance assay
Surface plasmon resonance assay was performed using a BIAcore 3000 instrument. GST-tagged K1D fragment of KAHRP (1 μM) was covalently coupled to a CM-5 biosensor chip using an amino coupling kit. Binding reactions were done in HBS-EP buffer, containing 20 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, and 0.05% (v/v) surfactant P20. The surface was regenerated before each new injection using 50 mM NaOH. The BIAcore instrument was programmed to perform a series of binding assays with increasing concentrations of MBP-MBD and MBP-D3 polypeptide over the same regenerated surface. Sensograms (plots of changes in response unit on the surface as a function of time) derived were analyzed using the software BIAeval 3.0. Affinity constants were estimated by curve fitting using a 1:1 binding model.
2.7. Resealing of erythrocytes
To introduce MBP, or MBP-tagged D3 into erythrocytes, the washed erythrocytes were lysed and resealed using the dialysis method [14,27]. Briefly, 2 ml of packed erythrocytes (in a dialysis tube, in the absence or presence of desired polypeptides) at a 50% hematocrit was dialyzed against 500 ml of cold hypotonic buffer (5 mM KPO4, pH 7.4, 20 mM KCl) for 80 min at 4 °C. The isotonicity was restored, and erythrocytes were resealed by dialyzing the lysed cells against 500 ml of pre-warmed isotonic buffer (5 mM KPO4, 160 mM KCl, 5 mM glucose) for 60 min at 37 °C.
2.8. Invasion of erythrocytes by malaria parasites
The 3D7 clone parasite was maintained using standard procedures in RPMI 1640 medium [28]. Sorbitol synchronization of P. falciparum-infected erythrocytes was performed as described before [29]. The synchronized infected cells were purified using Percoll gradients to nearly 100% purity [30]. The fraction containing mature trophozoite-infected erythrocytes was added to resealed erythrocytes at a starting parasitemia of 2%. The cells were cultured using standard procedures in RPMI 1640 medium for 48 h. At the end of the culture, the infected erythrocytes were separated by Percoll gradient, and the purified late stage infected cells were examined by confocal microscopy.
2.9. Immunofluorescence staining and confocal microscopy
Preparation of erythrocytes for immunofluorescence was performed as previously described [16,31]. Cells were permeabilized in PBS containing 0.1 M glycine (rinsing buffer) and 0.1% Triton-X100 for 5 min followed by three rinses with rinsing buffer. Nonspecific binding was blocked by incubation for at least 60 min in blocking buffer (PBS containing 0.05 mM glycine, 0.2% fish skin gelatin, and 0.05% sodium azide). Staining of fixed, permeabilized cells was performed using an anti-KAHRP antibody diluted in blocking buffer. After labeling, cells were re-suspended in PBS and allowed to attach to cover slips coated with polylysine. The cover slips were mounted using Aqua-Mount medium. Samples were imaged with a Zeiss 510 META confocal microscope using a 100× oil objective.
2.10. SDS-PAGE and Western blot
SDS-PAGE was performed in 10% acrylamide gels. Proteins were transferred onto nitrocellulose membrane. After blocking for 1 h in blocking buffer (10 mM Tris, pH 7.4, 150 mM NaCl, 0.5% Tween 20, 5% nonfat dry milk), the blot was probed for 1 h with the desired primary antibody (polyclonal anti-ankyrin R or anti-GST). After several washes, the blot was incubated with an anti-rabbit IgG, coupled to horseradish peroxidase (HRP). After final washes, the blot was developed using the Renaissance chemiluminescence detection kit. All steps were performed at room temperature.
3. Results
3.1. Association of malaria proteins with ankyrin R
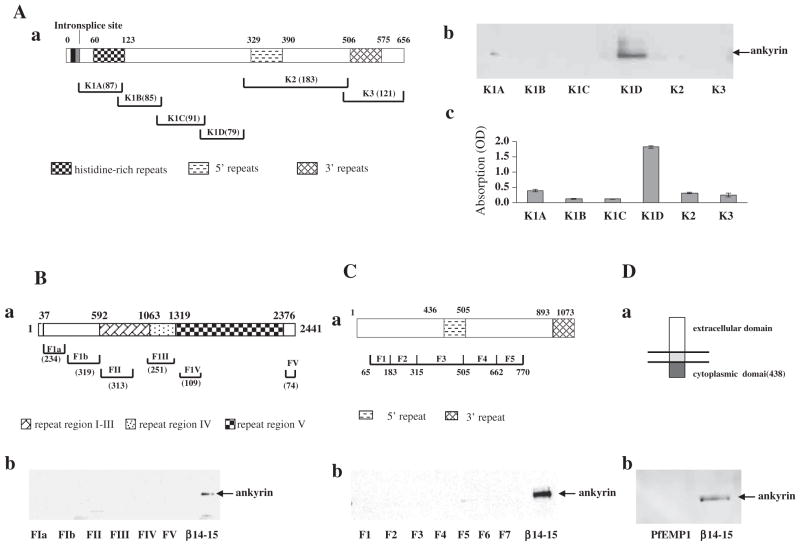
We first examined the binding of full length ankyrin R to fragments of the exported malarial proteins, PfEMP3, PfEMP1, RESA, and KAHRP using pulldown assays. All the recombinant polypeptides of above mentioned malaria proteins were well characterized and used in our previous published studies [14–16,24,25]. As a positive control, we used repeats 14–15 of β-spectrin, a well-established binder of ankyrin R [32]. A schematic representation of KAHRP structure and the recombinant fragments used in the present study is illustrated in Fig. 1A–a. As shown in Fig. 1A–b, of the six GST-tagged KAHRP fragments (K1A, K1B, K1C, K1D, K2, and K3), which encompass the full-length of KAHRP, only GST-K1D pulled down ankyrin R. The identification of this one fragment was further confirmed by ELISA assay with immobilized ankyrin R (Fig. 1A–c). Ankyrin R did not bind to any of the fragments of PfEMP3 (Fig. 1B–b), RESA (Fig. 1C–b) or the cytoplasmic domain of PfEMP1 (Fig. 1D–b). The positive control, ankyrin R bound to repeats 14–15 of spectrin. Thus ankyrin R is a binding partner for KAHRP on the erythrocyte membrane, and the binding site lies within the K1D region.
Fig. 1.
Association of malaria proteins with ankyrin R. (A) Binding of ankyrin R to KAHRP fragments. Schematic representation of KAHRP (a). The binding between ankyrin R and KAHRP fragments was assessed by pull-down (b) and ELISA assay (c). Note that ankyrin R bound to only the K1D fragment of the KAHRP. (B) Binding of ankyrin R to PfEMP3 fragments. Schematic representation of PfEMP3 (a). The binding between ankyrin R and PfEMP3 fragments was assessed by pull-down (b). Repeats 14–15 fragment of β-spectrin was used as a positive control for ankyrin R binding. (C) Binding of ankyrin R to RESA fragments. Schematic representation of RESA (a). The binding between ankyrin R and RESA fragments was assessed by pull-down (b). Repeats 14–15 fragment of β-spectrin was used as a positive control for ankyrin R binding. (D) Binding of ankyrin R to the cytoplasmic domain of PfEMP1. Schematic representation of PfEMP1 (a). The binding between ankyrin R and cytoplasmic domain of PfEMP1 was assessed by pull-down (b). Repeats 14–15 fragment of β-spectrin was used as a positive control for ankyrin R binding. Amino acid residue numbers and the lengths of fragments are indicated.
3.2. Mapping the KAHRP binding site in ankyrin R
A schematic representation of ankyrin R structure [33] and the recombinant fragments used in the present study are illustrated in Fig. 2A. We used a similar pull-down approach to map the binding site in this protein for KAHRP. Three recombinant MBP-tagged ankyrin R domains (the 89 kDa membrane-binding domain (MBD), the spectrin-binding domain and the C-terminal regulatory domain), which encompass full-length ankyrin R, were purified and examined for their ability to bind the K1D using both the MBP-pull-down assay and ELISA. As shown in Fig. 2B–a, under the binding conditions used in these experiments only the MBD fragment of ankyrin R was able to pull down K1D. K1D did not bind to either of the other two domains of ankyrin R. The selective binding of K1D to MBD is further confirmed by the ELISA assay which confirms that only K1D binds to immobilized MBD (Fig. 2B–b). Ankyrin R MBD domain is composed of four subdomains termed D1, D2, D3 and D4. To further define the KAHRP binding site in ankyrin R, the binding of GST-tagged K1D to MBP-tagged D1, D2, D3, and D4 was examined. Fig. 2C–a shows that only the D3 subdomain of ankyrin R was able to pull down K1D. Consistent with this finding, as shown in Fig. 2C–b K1D binds only to immobilized D3 by ELISA. Thus, we localized the KAHRP binding site in ankyrin R to the D3 region within MBD of ankyrin R.
Fig. 2.
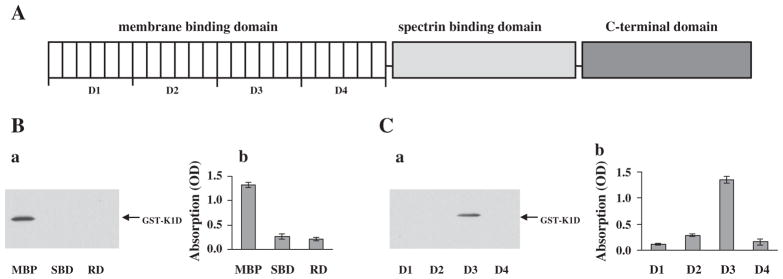
Binding site of KAHRP in ankyrin R. (A) Schematic representation of ankyrin R functional domains. (B) Binding of K1D to functional domains of ankyrin R. The binding of GST-tagged K1D fragment to MBP-tagged ankyrin R domains was assessed by MBP pull-down assay (left panel) and ELISA (right). Note that K1D bound to only the MBD of ankyrin R. (C) Binding of K1D fragment to subdomains of the MBD domain of ankyrin R. The binding of GST-tagged K1D fragment to sub-domains of MBD was assessed by MBP pull-down assay (a) and by ELISA assay (b). Note that K1D bound to only the D3 region of the MBD domain of ankyrin R.
3.3. Kinetic analysis of interactions between KAHRP and ankyrin R by surface plasmon resonance assay
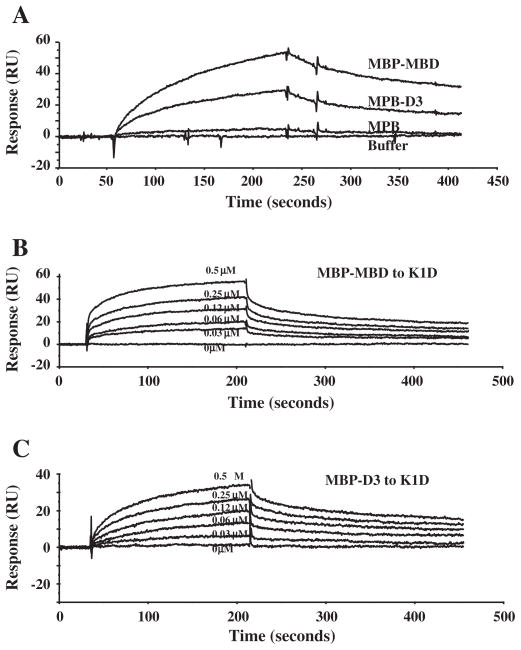
We used the active binding fragments determined above to obtain kinetic binding data by surface plasmon resonance assay. In these experiments, the GST-tagged K1D fragment of KAHRP was immobilized on the sensor chip and the binding of ankyrin R MBD, or of its D3 subdomain was followed. As shown in Fig. 3, MBP-tagged MBD and D3, but not MBP alone, bound to the immobilized K1D (Fig. 3A). Fig. 3B and C demonstrates the dose-dependent binding of MBD and D3 to K1D respectively. Curve fitting of the sensograms enabled us to derive the association and dissociation constants and derive the binding affinity values (Table 1). The binding affinity for MBD–K1D or D3–K1D interaction is 3.8 × 10−8 M and 4.6 × 10−8 M respectively.
Fig. 3.
Kinetic analysis of interaction between K1D and ankyrin R polypeptides by surface plasmon resonance assay. A: GST-tagged K1D fragment was immobilized onto a CM5 sensor chip. MBP, MBP-MBD or MBP-D3 at a concentration of 1 μM was injected at 20 μl/min over the chip surface in a BIAcore 3000 instrument. B: MBP-MBD at different concentrations as indicated was injected at 20 μl/min over the surface in a BIAcore 3000 instrument. The figure shows dose–response curves of MBP-MBD binding to K1D. C: MBP-D3 at different concentrations as indicated was injected at 20 μl/min over the surface in a BIAcore 3000 instrument. The figure shows dose–response curves of MBP-D3 binding to K1D.
Table 1.
Binding affinities between ankyrin R fragments to KAHRP fragment.
| Ka(×104M−1 s−1) | Kd(×10−3 s−1) | KD(×10−8 M) | |
|---|---|---|---|
| MBD–K1D | 4.28 | 1.61 | 3.8 |
| D3–K1D | 2.99 | 1.37 | 4.6 |
3.4. Inhibition of ankyrin R binding to KAHRP by an ankyrin R fragment
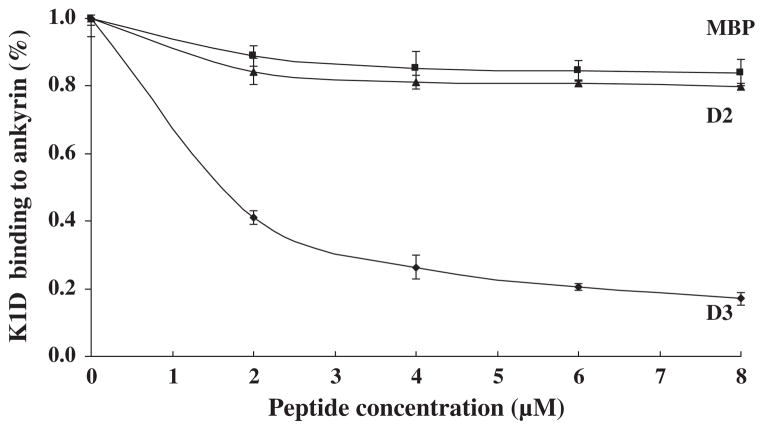
To confirm the specificity of the interaction between ankyrin R and KAHRP, a competitive inhibition assay was performed. The K1D fragment was incubated with varying concentrations of D3 before adding it to a 96-well plate coated with ankyrin R. As shown in Fig. 4, the binding of K1D to the intact protein decreased with increasing concentrations of ankyrin R D3 fragment. At a concentration of 8 μM of D3, the binding between ankyrin R and KAHRP was inhibited by 80%. MBP and MBP-tagged D2 fragments of ankyrin R had no inhibitory effect on the binding of the K1D fragment to ankyrin R.
Fig. 4.
Inhibition of binding of K1D to ankyrin R by the D3 fragment of ankyrin R. K1D was incubated with increasing concentrations of D3 or MBP before addition to immobilized ankyrin R in the ELISA wells. K1D binding to ankyrin R is inhibited by D3.
3.5. Effect of the ankyrin R D3 fragment on KAHRP localization in malaria-infected erythrocytes
We have previously shown that the association of KAHRP with the host cytoskeleton proteins is required for its proper assembly on the erythrocyte membrane [16]. To establish the function of KAHRP–ankyrin R interaction, we examined the localization of KAHRP in erythrocytes, pre-loaded with free D3 ankyrin R fragment, and then exposed to invasion by P. falciparum. Fig. 5A shows the membrane association of KAHRP in resealed control erythrocytes infected by the parasite. The same occurred when the resealed erythrocytes contained MBP (Fig. 5B). In marked contrast, in cells containing MBP-D3 the KAHRP showed only a diffuse distribution throughout the cytoplasm (Fig. 5C). Uninfected erythrocytes showed no staining with an anti-KAHRP antibody (data not shown). Thus sequestration of KAHRP by its free ankyrin R-binding fragment prevents binding of the KAHRP to the host cell membrane. We are aware that MBP is a large fusion tag which could cause steric interference, but the finding that resealing of MBP itself had no effect on the localization of KAHRP strongly suggests that the altered localization of KAHRP in MBP-D3 resealed cells is due to D3 but not MBP.
Fig. 5.
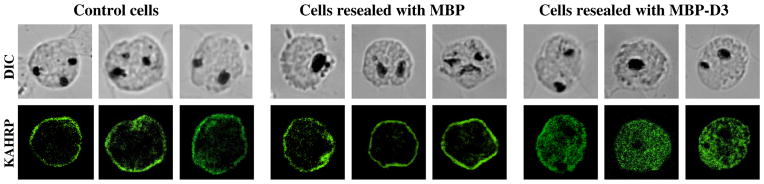
Localization and distribution of KAHRP in erythrocytes infected with P. falciparum. Erythrocytes were resealed with no added proteins (A), with 40 μM MBP (B), or with 40 μM MBP-D3 (C). The resealed erythrocytes were infected with P. falciparum and cultured for 48 h. The localization of KAHRP was detected by immunostaining with an anti-KAHRP antibody (green color). The stained cells were imaged by confocal microscopy (100× objection).
4. Discussion
In the present study, we performed a detailed molecular characterization of the interaction between KAHRP and ankyrin R. We showed that the association of the malaria protein KAHRP with the host cytoskeleton protein ankyrin R is required for the attachment of KAHRP to the red cell membrane. The importance of ankyrin R in malaria infection is heighted by recent findings that the growth of malaria parasite was impaired in ankyrin R mutant red cells [34,35]. Although the mechanisms for the parasite growth retardation in ankyrin R mutant red cells remain largely unknown, it is possible that the defective KAHRP–ankyrin R association in these cells may contribute to this.
We have previously shown that KAHRP binds to α-spectrin repeat 4 [16], and this part of α-spectrin is close to the spectrin tetramerization site. As ankyrin R binds to repeats 14–15 of β-spectrin which is also close to the spectrin tetramerization site [32], this arrangement suggests that it is possible for a single molecule of KAHRP to engage in stabilizing two reactions at the membrane skeleton, one with ankyrin R and one with spectrin. It should be noted that while the binding affinity between KAHRP and α-spectrin is in the micromolar range [16], the binding affinity between KAHRP and ankyrin R is in the subnanomolar range, suggesting that KAHRP association with the membrane in infected erythrocytes may be dominantly regulated by ankyrin R.
Our earlier work also suggested an interaction between KAHRP and ankyrin R, however it was based on indirect experiments in which solubilized erythrocyte proteins were immunoprecipitated with an anti-KAHRP antibody [23]. Such studies may identify spurious interactions by bringing together proteins that are not normally associated. In that study we also used recombinant fragments in an attempt to map interacting protein domains. The level of resolution possible at the time was less than is available presently, nevertheless, we did suggest that the 89 kDa MBD of ankyrin R was involved in the interaction with KAHRP. In our present study, using multiple independent experimental approaches we confirm that KAHRP and ankyrin R do indeed interact and that this interaction resides within the MBD of ankyrin R. One discrepancy between the two studies is that we previously assigned the ankyrin R binding domain to the repeat region of the K2 fragment of KAHRP, a different conclusion from the present study which assigns this binding domain to K1D. We have no explanation for this discrepancy but we note that the affinity of the previously identified interaction was 20-fold less than the affinity measured in the current study. Furthermore, in contrast to the earlier study we performed an extensive series of studies to document the specificity of the interaction between K1D fragment of KAHRP and ankyrin R.
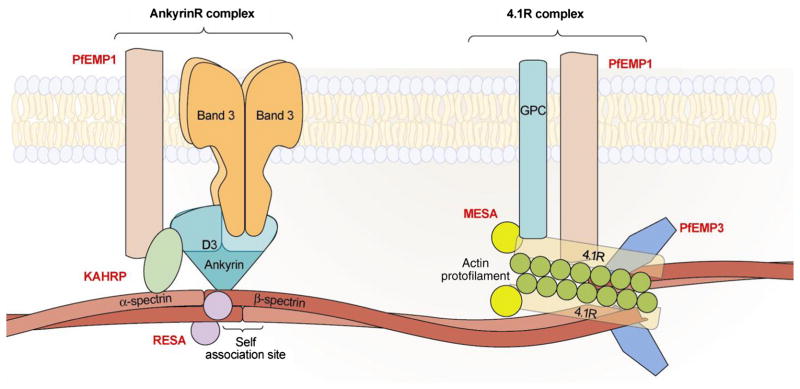
KAHRP is a major component of the knob, an electron-dense protrusion located at the membrane of infected erythrocytes, the site of adhesive interaction between infected erythrocytes and vascular endothelial cells [36]. Although PfEMP1, the adhesive receptor expressed on the surface of infected erythrocytes is responsible for adhesive interactions, KAHRP by clustering PfEMP1 molecules markedly increases the avidity of these adhesive interactions. Strong support for such a thesis comes from the finding that that in the absence of KAHRP, infected erythrocytes adhere very weakly to endothelial cells under physiological flow conditions [37]. It has previously been suggested that PfEMP1 is anchored to the junctional complex through its direct interaction with spectrin and actin or by its association with KAHRP [38,39]. However, because the KAHRP binding site in spectrin is remote from the spectrin–actin–protein 4.1R junctional complex and because KAHRP does not bind to 4.1R (our unpublished data), we suggest that the direct association of PfEMP1 and KAHRP probably occurs at the ankyrin R complex rather than at the junctional complex. We further suggest that PfEMP1 may be anchored to the membrane skeleton at both ankyrin R complex and 4.1R-complex. Based on the previous findings and the findings from the present study, we propose the current schematic model for the interactions between malaria proteins and host skeleton proteins in the infected erythrocytes. Fig. 6 summarizes the interactions. At the ankyrin R-based macromolecular complex (which is close to spectrin tetramerization site), PfEMP1 binds to KAHRP which in turn binds to both ankyrin R and repeat 4 of α-spectrin [16]. RESA binds to repeat 16 of β-spectrin and strengthens the spectrin tetramer formation [14]. At the 4.1R-based macromolecular complex, PfEMP1 is anchored to the junctional complex through its direct binding to spectrin, actin and 4.1R; PfEMP3 binds to the EF hand domains of α-spectrin and interferes with the spectrin–actin–4.1R ternary complex formation and destabilizes the membrane stability [15]; MESA binds to the 30 kDa membrane domain of 4.1R [19]. It is interesting to note that all identified associations centered around either ankyrin R-based macromolecular complex or 4.1R-based macromolecular complex.
Fig. 6.
Schematic representation of interactions between malaria proteins and host skeleton proteins in malaria infected erythrocytes. At the ankyrin R-based macromolecular complex, which is near the center of the tetramer (dimer–dimer interaction site), PfEMP1 binds to KAHRP which in turn binds to both ankyrin R and α-spectrin. RESA binds to repeat 16 of β-spectrin. At the 4.1R-based macromolecular complex, PfEMP1 is anchored to the junctional complex through its direct binding to spectrin, actin and 4.1R. PfEMP3 also directly binds to spectrin, actin and 4.1R. MESA binds to the 30 kDa membrane domain of 4.1R.
We have previously documented that spectrin plays an important role in localizing KAHRP to the erythrocyte membrane. In the present study, we show that ankyrin R plays a similar role. It appears that both interactions are required as each can be separately inhibited by the addition of an excess of ligand and in each case KAHRP mislocalizes. As KAHRP is important for adherence of infected erythrocytes and adherence of infected erythrocytes to vascular endothelium is a major determinant of the pathogenicity of P. falciparum [40], we suggest that reducing or eliminating adhesive interactions by interfering with the interaction of KAHRP with spectrin and ankyrin R is a potential therapeutic strategy against malaria infection.
We and others have documented that the majority of characterized associations between malaria proteins and the membrane of infected red cells are mediated via interaction with spectrin. Unexpectedly, in the present study we found that of the four spectrin-binding malaria proteins (PfEMP1, PfEMP3, RESA and KAHRP), only KAHRP binds to ankyrin R. These results suggest that spectrin is the major substrate for localization of exported malarial proteins to the red cell membrane skeleton and that spectrin may serve as a scaffold for assembly of malaria proteins in infected erythrocytes on the red cell membrane. However, it should be noted that we could not exclude the possibility that the lack of association of other malaria proteins with ankyrin R may be due to some interactions require the cooperation of two domains being co-expressed rather than one.
Previous studies have identified binding motifs involved in the interactions between malaria proteins and erythrocyte membrane proteins. For example, the 4.1R-binding site in MESA has been localized to 19 residues located in the N-terminal region of MESA [18] and a recent study further defined similar erythrocyte cytoskeleton-binding motif in a number of exported P. falciparum proteins including MESA [41]. The spectrin-binding domain in RESA has been mapped to a 48-residue region [42], whereas the spectrin-binding domain in MSP-1 has been localized to a 30-residue region [43]. We have identified the reciprocal binding sites for malaria proteins on erythrocyte proteins. For example, the MESA binding site on 4.1R has been mapped to a 51-residue region encoded by exon 10 of the 4.1R gene [19]. The KAHRP, PfEMP3 and RESA binding sites on spectrin have been mapped to repeat 4 of α-spectrin, the EF hand domains of α-spectrin and repeat 16 of β-spectrin respectively [14–16]. It is generally believed that the associations between a group of malaria proteins and erythrocyte proteins are responsible for almost all the altered properties of parasitized erythrocytes. It should be noted that according to proteomic analyses there are at least 400 exported blood-stage P. falciparum proteins [1–3]. Thus identification and characterization of these associations are still far from complete. All of these interactions appear to be highly specific and in at least some cases result in defined alterations in mechanical properties of infected cells. These studies have been performed in isolation from each other and we still await findings from future studies that examine the simultaneous effects of multiple interacting domains on red cell function.
Footnotes
This work was supported in part by the NIH grants DK 26263 and DK 32094, and by grants 81171905 and 81272187 from the Natural Science Foundation of China.
References
- 1.Hiller NL, Bhattacharjee S, van Ooij C, Liolios K, Harrison T, et al. A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science. 2004;306:1934–1937. doi: 10.1126/science.1102737. [DOI] [PubMed] [Google Scholar]
- 2.Marti M, Baum J, Rug M, Tilley L, Cowman AF. Signal-mediated export of proteins from the malaria parasite to the host erythrocyte. J Cell Biol. 2005;171:587–592. doi: 10.1083/jcb.200508051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sargeant TJ, Marti M, Caler E, Carlton JM, Simpson K, et al. Lineage-specific expansion of proteins exported to erythrocytes in malaria parasites. Genome Biol. 2006;7:R12. doi: 10.1186/gb-2006-7-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oh SS, Chishti AH, Palek J, Liu SC. Erythrocyte membrane alterations in Plasmodium falciparum malaria sequestration. Curr Opin Hematol. 1997;4:148–154. doi: 10.1097/00062752-199704020-00012. [DOI] [PubMed] [Google Scholar]
- 5.Maier AG, Cooke BM, Cowman AF, Tilley L. Malaria parasite proteins that remodel the host erythrocyte. Nat Rev Microbiol. 2009;7:341–354. doi: 10.1038/nrmicro2110. [DOI] [PubMed] [Google Scholar]
- 6.Cooke BM, Mohandas N, Coppel RL. The malaria-infected red blood cell: structural and functional changes. Adv Parasitol. 2001;50:1–86. doi: 10.1016/S0065-308X(01)50029-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennett V. Spectrin-based membrane skeleton: a multipotential adaptor between plasma membrane and cytoplasm. Physiol Rev. 1990;70:1029–1065. doi: 10.1152/physrev.1990.70.4.1029. [DOI] [PubMed] [Google Scholar]
- 8.Mohandas N, Gallagher PG. Red cell membrane: past, present, and future. Blood. 2008;112:3939–3948. doi: 10.1182/blood-2008-07-161166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.An X, Mohandas N. Disorders of red cell membrane. Br J Haematol. 2008;141:367–375. doi: 10.1111/j.1365-2141.2008.07091.x. [DOI] [PubMed] [Google Scholar]
- 10.De Matteis MA, Morrow JS. Spectrin tethers and mesh in the biosynthetic pathway. J Cell Sci. 2000;113(Pt 13):2331–2343. doi: 10.1242/jcs.113.13.2331. [DOI] [PubMed] [Google Scholar]
- 11.Bennett V, Baines AJ. Spectrin and ankyrin-based pathways: metazoan inventions for integrating cells into tissues. Physiol Rev. 2001;81:1353–1392. doi: 10.1152/physrev.2001.81.3.1353. [DOI] [PubMed] [Google Scholar]
- 12.Bruce LJ, Beckmann R, Ribeiro ML, Peters LL, Chasis JA, et al. A band 3-based macrocomplex of integral and peripheral proteins in the RBC membrane. Blood. 2003;101:4180–4188. doi: 10.1182/blood-2002-09-2824. [DOI] [PubMed] [Google Scholar]
- 13.Salomao M, Zhang X, Yang Y, Lee S, Hartwig JH, et al. Protein 4.1R-dependent multiprotein complex: new insights into the structural organization of the red blood cell membrane. Proc Natl Acad Sci U S A. 2008;105:8026–8031. doi: 10.1073/pnas.0803225105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pei X, Guo X, Coppel R, Bhattacharjee S, Haldar K, et al. The ring-infected erythrocyte surface antigen (RESA) of Plasmodium falciparum stabilizes spectrin tetramers and suppresses further invasion. Blood. 2007;110:1036–1042. doi: 10.1182/blood-2007-02-076919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pei X, Guo X, Coppel R, Mohandas N, An X. Plasmodium falciparum erythrocyte membrane protein 3 (PfEMP3) destabilizes erythrocyte membrane skeleton. J Biol Chem. 2007;282:26754–26758. doi: 10.1074/jbc.M701612200. [DOI] [PubMed] [Google Scholar]
- 16.Pei X, An X, Guo X, Tarnawski M, Coppel R, et al. Structural and functional studies of interaction between Plasmodium falciparum knob-associated histidine-rich protein (KAHRP) and erythrocyte spectrin. J Biol Chem. 2005;280:31166–31171. doi: 10.1074/jbc.M505298200. [DOI] [PubMed] [Google Scholar]
- 17.Lustigman S, Anders RF, Brown GV, Coppel RL. The mature-parasite-infected erythrocyte surface antigen (MESA) of Plasmodium falciparum associates with the erythrocyte membrane skeletal protein, band 4.1. Mol Biochem Parasitol. 1990;38:261–270. doi: 10.1016/0166-6851(90)90029-l. [DOI] [PubMed] [Google Scholar]
- 18.Bennett BJ, Mohandas N, Coppel RL. Defining the minimal domain of the Plasmodium falciparum protein MESA involved in the interaction with the red cell membrane skeletal protein 4.1. J Biol Chem. 1997;272:15299–15306. doi: 10.1074/jbc.272.24.15299. [DOI] [PubMed] [Google Scholar]
- 19.Waller KL, Nunomura W, An X, Cooke BM, Mohandas N, et al. Mature parasite-infected erythrocyte surface antigen (MESA) of Plasmodium falciparum binds to the 30-kDa domain of protein 4.1 in malaria-infected red blood cells. Blood. 2003;102:1911–1914. doi: 10.1182/blood-2002-11-3513. [DOI] [PubMed] [Google Scholar]
- 20.Black CG, Proellocks NI, Kats LM, Cooke BM, Mohandas N, et al. In vivo studies support the role of trafficking and cytoskeletal-binding motifs in the interaction of MESA with the membrane skeleton of Plasmodium falciparum-infected red blood cells. Mol Biochem Parasitol. 2008;160:143–147. doi: 10.1016/j.molbiopara.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magowan C, Coppel RL, Lau AO, Moronne MM, Tchernia G, et al. Role of the Plasmodium falciparum mature-parasite-infected erythrocyte surface antigen (MESA/PfEMP-2) in malarial infection of erythrocytes. Blood. 1995;86:3196–3204. [PubMed] [Google Scholar]
- 22.Chishti AH, Palek J, Fisher D, Maalouf GJ, Liu SC. Reduced invasion and growth of Plasmodium falciparum into elliptocytic red blood cells with a combined deficiency of protein 4.1, glycophorin C, and p55. Blood. 1996;87:3462–3469. [PubMed] [Google Scholar]
- 23.Magowan C, Nunomura W, Waller KL, Yeung J, Liang J, et al. Plasmodium falciparum histidine-rich protein 1 associates with the band 3 binding domain of ankyrin in the infected red cell membrane. Biochim Biophys Acta. 2000;1502:461–470. doi: 10.1016/s0925-4439(00)00069-7. [DOI] [PubMed] [Google Scholar]
- 24.Waller KL, Cooke BM, Nunomura W, Mohandas N, Coppel RL. Mapping the binding domains involved in the interaction between the Plasmodium falciparum knob-associated histidine-rich protein (KAHRP) and the cytoadherence ligand P. falciparum erythrocyte membrane protein 1 (PfEMP1) J Biol Chem. 1999;274:23808–23813. doi: 10.1074/jbc.274.34.23808. [DOI] [PubMed] [Google Scholar]
- 25.Waller KL, Stubberfield LM, Dubljevic V, Nunomura W, An X, et al. Interactions of Plasmodium falciparum erythrocyte membrane protein 3 with the red blood cell membrane skeleton. Biochim Biophys Acta. 2007;1768:2145–2156. doi: 10.1016/j.bbamem.2007.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tyler JM, Hargreaves WR, Branton D. Purification of two spectrin-binding proteins: biochemical and electron microscopic evidence for site-specific reassociation between spectrin and bands 2.1 and 4.1. Proc Natl Acad Sci U S A. 1979;76:5192–5196. doi: 10.1073/pnas.76.10.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dluzewski AR, Rangachari K, Wilson RJ, Gratzer WB. Properties of red cell ghost preparations susceptible to invasion by malaria parasites. Parasitology. 1983;87(Pt 3):429–438. doi: 10.1017/s0031182000082950. [DOI] [PubMed] [Google Scholar]
- 28.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 29.Aley SB, Sherwood JA, Marsh K, Eidelman O, Howard RJ. Identification of isolate-specific proteins on sorbitol-enriched Plasmodium falciparum infected erythrocytes from Gambian patients. Parasitology. 1986;92(Pt 3):511–525. doi: 10.1017/s0031182000065410. [DOI] [PubMed] [Google Scholar]
- 30.Aley SB, Sherwood JA, Howard RJ. Knob-positive and knob-negative Plasmodium falciparum differ in expression of a strain-specific malarial antigen on the surface of infected erythrocytes. J Exp Med. 1984;160:1585–1590. doi: 10.1084/jem.160.5.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campanella ME, Chu H, Low PS. Assembly and regulation of a glycolytic enzyme complex on the human erythrocyte membrane. Proc Natl Acad Sci U S A. 2005;102:2402–2407. doi: 10.1073/pnas.0409741102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kennedy SP, Warren SL, Forget BG, Morrow JS. Ankyrin binds to the 15th repetitive unit of erythroid and nonerythroid beta-spectrin. J Cell Biol. 1991;115:267–277. doi: 10.1083/jcb.115.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michaely P, Bennett V. The membrane-binding domain of ankyrin contains four independently folded subdomains, each comprised of six ankyrin repeats. J Biol Chem. 1993;268:22703–22709. [PubMed] [Google Scholar]
- 34.Rank G, Sutton R, Marshall V, Lundie RJ, Caddy J, et al. Novel roles for erythroid ankyrin-1 revealed through an ENU-induced null mouse mutant. Blood. 2009;113:3352–3362. doi: 10.1182/blood-2008-08-172841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greth A, Lampkin S, Mayura-Guru P, Rodda F, Drysdale K, et al. A novel ENU-mutation in ankyrin-1 disrupts malaria parasite maturation in red blood cells of mice. PLoS One. 2012;7:e38999. doi: 10.1371/journal.pone.0038999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berendt AR, Ferguson DJ, Gardner J, Turner G, Rowe A, et al. Molecular mechanisms of sequestration in malaria. Parasitology. 1994;108:S19–S28(Suppl). doi: 10.1017/s0031182000075685. [DOI] [PubMed] [Google Scholar]
- 37.Crabb BS, Cooke BM, Reeder JC, Waller RF, Caruana SR, et al. Targeted gene disruption shows that knobs enable malaria-infected red cells to cytoadhere under physiological shear stress. Cell. 1997;89:287–296. doi: 10.1016/s0092-8674(00)80207-x. [DOI] [PubMed] [Google Scholar]
- 38.Oh SS, Voigt S, Fisher D, Yi SJ, LeRoy PJ, et al. Plasmodium falciparum erythrocyte membrane protein 1 is anchored to the actin-spectrin junction and knob-associated histidine-rich protein in the erythrocyte skeleton. Mol Biochem Parasitol. 2000;108:237–247. doi: 10.1016/s0166-6851(00)00227-9. [DOI] [PubMed] [Google Scholar]
- 39.Voigt S, Hanspal M, LeRoy PJ, Zhao PS, Oh SS, et al. The cytoadherence ligand Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) binds to the P. falciparum knob-associated histidine-rich protein (KAHRP) by electrostatic interactions. Mol Biochem Parasitol. 2000;110:423–428. doi: 10.1016/s0166-6851(00)00281-4. [DOI] [PubMed] [Google Scholar]
- 40.MacPherson GG, Warrell MJ, White NJ, Looareesuwan S, Warrell DA. Human cerebral malaria. A quantitative ultrastructural analysis of parasitized erythrocyte sequestration. Am J Pathol. 1985;119:385–401. [PMC free article] [PubMed] [Google Scholar]
- 41.Kilili GK, LaCount DJ. An erythrocyte cytoskeleton-binding motif in exported Plasmodium falciparum proteins. Eukaryot Cell. 2011;10:1439–1447. doi: 10.1128/EC.05180-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Foley M, Corcoran L, Tilley L, Anders R. Plasmodium falciparum: mapping the membrane-binding domain in the ring-infected erythrocyte surface antigen. Exp Parasitol. 1994;79:340–350. doi: 10.1006/expr.1994.1096. [DOI] [PubMed] [Google Scholar]
- 43.Herrera S, Rudin W, Herrera M, Clavijo P, Mancilla L, et al. A conserved region of the MSP-1 surface protein of Plasmodium falciparum contains a recognition sequence for erythrocyte spectrin. EMBO J. 1993;12:1607–1614. doi: 10.1002/j.1460-2075.1993.tb05805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]