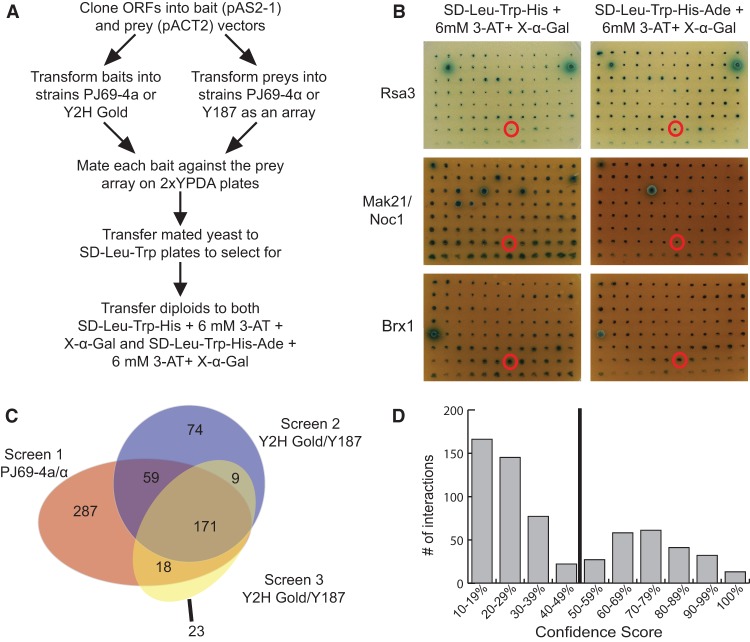
Figure 1.
An array based Y2H screen identifies novel interactions among LSU processome proteins. (A) Work flow of the array-based Y2H screen. (B) Results from Y2H screen 2 performed in Y2H Gold/Y187 for Rsa3, Mak21/Noc1, and Brx1 bait proteins. Each bait was assayed against all 70 arrayed preys on two different selective media: SD−Leu−Trp−His + 6 mM 3-amino-1,2,4-triazole (3-AT) + 40 μg/mL 5-bromo-4-chloro-3-indolyl α-D-galactopyranoside (X-α-Gal) or SD−Leu−Trp−His−Ade + 6 mM 3-AT + 40 μg/mL X-α-Gal. Growth of a blue colony on either selective medium is indicative of a positive Y2H interaction. (C) A Venn diagram summarizing the PPIs that were identified in each of the three iterations of the array-based Y2H screen. Screen 1 was performed in the PJ69-4a/α yeast strains, while screens 2 and 3 were performed in the Y2H Gold/Y187 yeast strains from Clontech. The number of interactions that were unique to each screen and the number of interactions that were identified in more than one screen are indicated. (D) Histogram of the confidence score distribution for the 641 PPIs identified in this study. The confidence score reflects the reproducibility of an observed interaction across all screens and selective media. Interactions with a confidence score >50% were included in the high-confidence LSU processome interactome data set and are listed in Supplemental Table S3.