Figure 5.
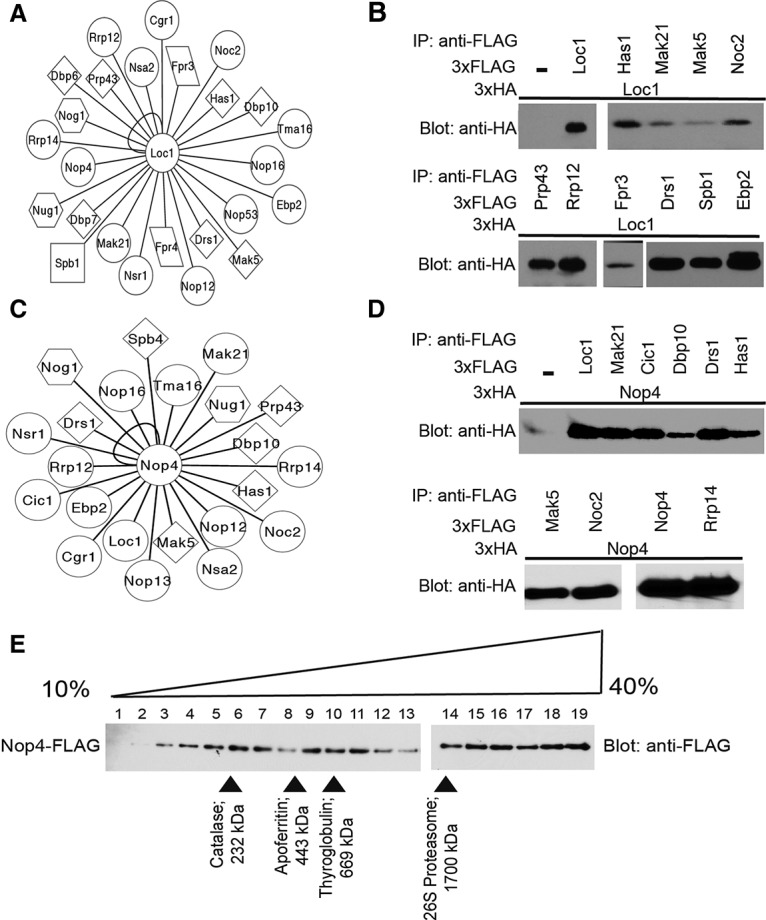
Loc1 and Nop4 are hub proteins within the LSU processome interactome. (A) Network of novel, high-confidence Y2H interactions identified for Loc1. Circles represent proteins with no known enzymatic activity, diamonds represent helicases, squares represent methyltransferases, hexagons represent GTPases, and parallelograms represent PPIases. The interaction network was created using Cytoscape. (B) Validation of the novel Loc1 interactions by coimmunoprecipitation. Yeast extract was incubated with α-Flag resin. Coimmunoprecipitations were assessed by α-HA Western blot. (C) Network of novel, high-confidence Y2H interactions identified for Nop4. Circles represent proteins with no known enzymatic activity, diamonds represent helicases, and hexagons represent GTPases. The interaction network was created using Cytoscape. (D) Validation of the novel Nop4 interactions by coimmunoprecipitation. Yeast extract was incubated with α-Flag resin. Coimmunoprecipitations were assessed by α-HA Western blot. (E) Glycerol gradient sedimentation analysis of Nop4. Yeast extract made from YPH499 Nop4-Flag was layered onto a 10%–40% glycerol gradient. The gradients were spun at 150,000g for 18 h, and fractions were harvested from the top of the gradient. All fractions were analyzed by α-Flag Western blot.
