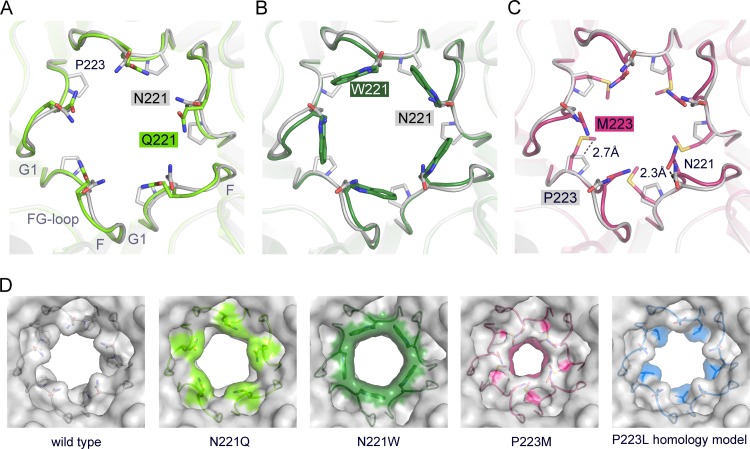
FIG 4.
Crystal structures of VP1 pentamers with N221Q, N221W, and P223M pore mutations. (A to C) Structural superposition of pore mutants N221Q (A), N221W (B), and P221M (C) with wild-type Mad-1 VP1. VP1 pentamers are shown in cartoon representations with side chains of key amino acid residues highlighted in stick representations and colored according to atom type (oxygens are shown in red, nitrogens in blue, and carbons in the colors assigned for the respective mutants). Wild-type VP1 is shown in gray. The accessible diameter at the narrowest constriction of the 5-fold pore was determined (28). The diameter was calculated to be 8.2 Å for wild-type VP1 and to be 7.1 Å and 8.6 Å for N221Q and N222W, respectively. The P223M mutation results in a constrained accessible pore diameter of 5.2 Å. (D) Surface representations of the 5-fold pore. Views are equivalent in all cases, and mutated amino acids are colored on the surface. A VP1 homology model for P223L was generated, and the most favorable rotamer conformation of Leu is shown in a surface representation using sticks.