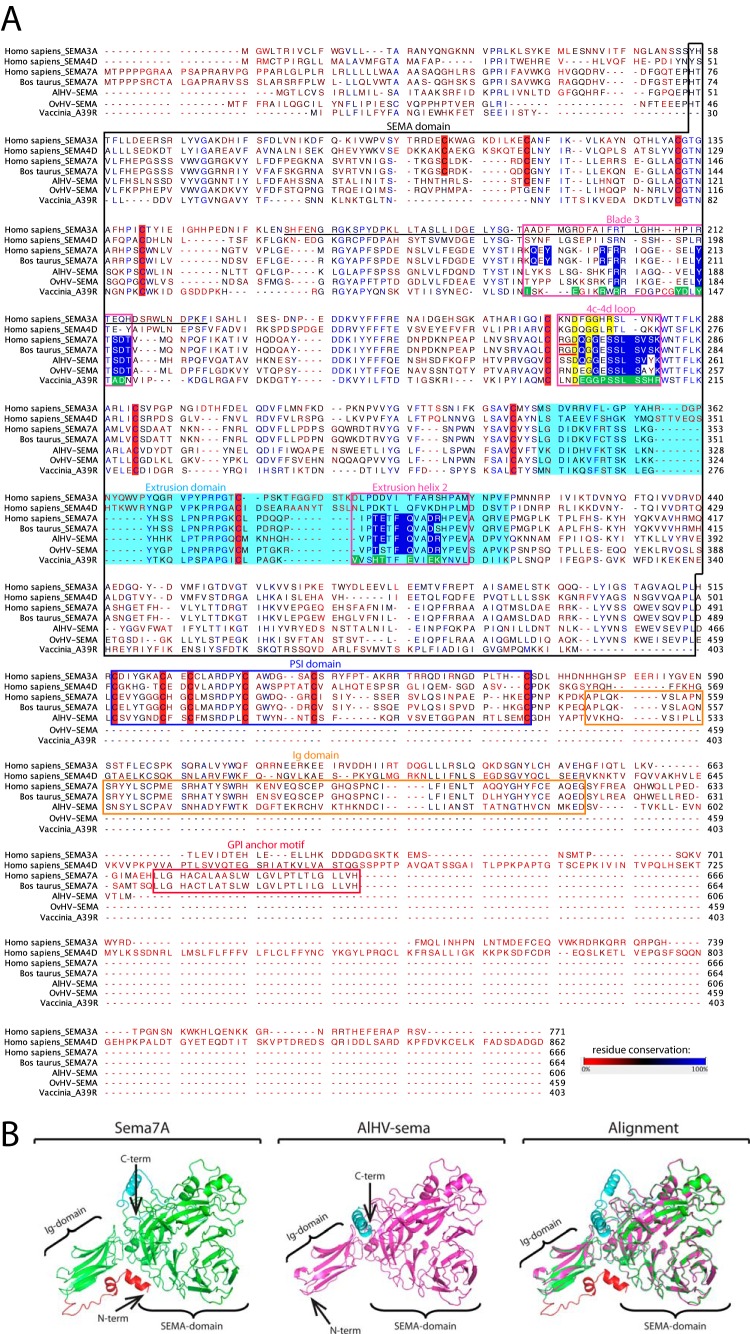
FIG 2.
Sequence alignment of human sema3A, sema4D, and sema7A, bovine sema7A and A39R, OvHV-sema, and AlHV-sema. (A) CLUSTALW was used to align peptidic sequences. Sequence analyses of sema7A and viral homologs were extrapolated from data available for other semaphorins (41). The SEMA domain is boxed in black. Extrusion and PSI domains are highlighted in light blue and boxed in blue, respectively. The 70-residue section of sema3A implicated in receptor specificity is underlined in black (53). The regions involved in the binding of sema7A and A39R to plexinC1 (blade 3, 4c-4d loop, and extrusion helix 2) are boxed in pink. Key residues involved in bonding interaction with plexinC1 are highlighted in blue for sema7A and macavirus semaphorins and in green for A39R (12). Conserved structural determinant in the 4c-4d loop are highlighted in yellow. RGD motives at the base of the sema7A 4c-4d loop are underlined in red. Cysteines conserved in the semaphorins are highlighted in red. The figure was produced with CLC Sequence Viewer v.7.0.2 and modified using Adobe Illustrator CS5. (B) 3-D prediction of the structure of AlHV-sema based on the crystal structure of human sema7A (12) (http://zhanglab.ccmb.med.umich.edu/I-TASSER/).