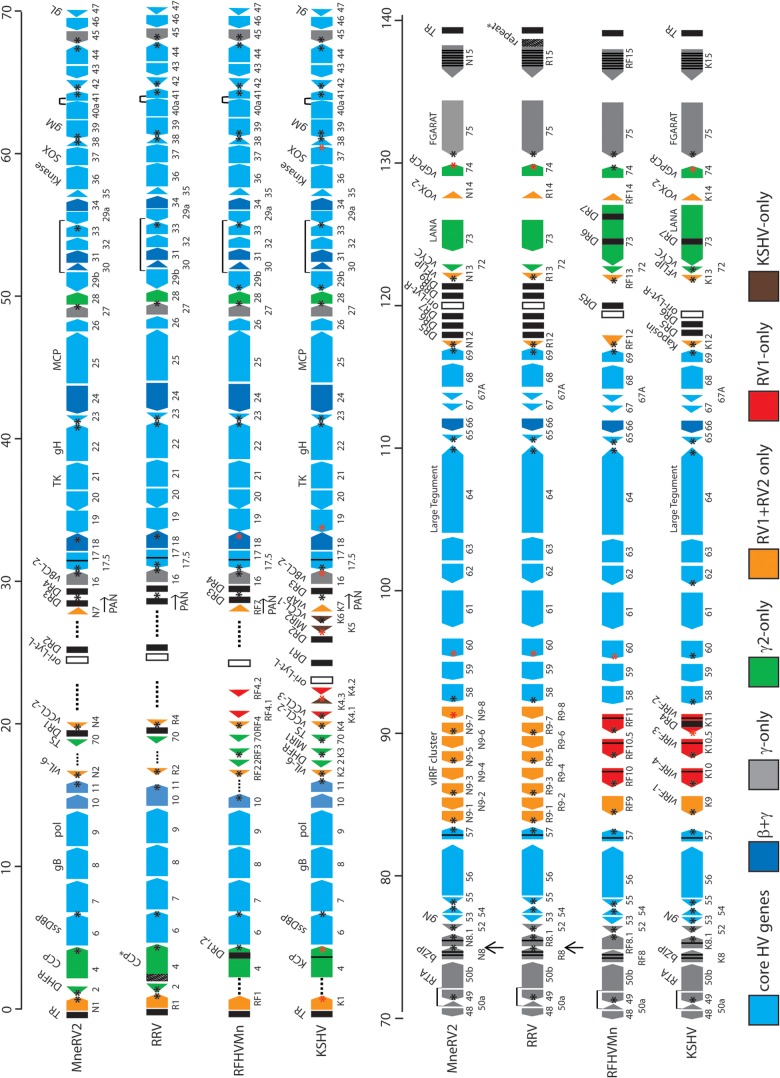
FIG 1.
Comparative map of the MneRV2, RRV, RFHVMn, and KSHV genomes. The positions and transcription directions of the ORFs identified in the MneRV2 genome are compared to the corresponding ORFs in RRV (strain 26-95 and strain 17577), RFHVMn, and KSHV (see Materials and Methods for accession records). A composite map of the two RRV strains is shown, as RRV17577 has a duplicated N-terminal domain in CCP/ORF4 (indicated as CCP*) while RRV26-95 has a large repeated domain disrupting the R15 homolog (indicated as R15-repeat*), as discussed in the text. The numbering of the MneRV2, RRV, RFHVMn, and KSHV ORFs is patterned after the genome structure of the prototype rhadinovirus, herpesvirus saimiri (HVS), with KSHV-specific ORFs designated K1 to K15 and their homologs in MneRV2, RRV, and RFHVMn designated with an N, R, or RF prefix, respectively. This follows the nomenclature convention established for the RRV26-95 and RFHVMn genome sequences. The 8-fold duplications of the K9/RF9 vIRF-1 gene found in the vIRF cluster in MneRV2 and both RRV strains are labeled as N9-1 to -8 and R9-1 to -8, respectively. The approximate positions of the ORFs are shown with regard to their position within the KSHV genome (i.e., 1 to 140 kb). Note the conserved location of the ORF2/DHFR gene upstream of ORF4 in MneRV2 and both RRV strains. The sizes of the ORF markers are approximately consistent with the sizes of the encoded proteins. Vertical black lines within an ORF indicate splicing events or internal initiations, while longer-range splices of protein coding exons are indicated with a bar. Transcription-terminating poly(A) signals identified in KSHV (42) and corresponding conserved signals in the other genomes are indicated as a black (AAUAAA) or red (AUUAAA) asterisk. The poly(A) signals downstream of MneRV2 and RRV ORFs N1/R1, N8/R8, 48, 60, and N15/R15, which are not present in KSHV or RFHVMn, as discussed in the text, are indicated with arrows. The ORFs are color coded with regard to their conservation in other herpesvirus genomes, showing core HV genes conserved in most herpesvirus families, both beta- and gammaherpesviruses, gammaherpesviruses only, gamma-2-herpesviruses only, RV1 and RV2 rhadinoviruses only, RV1 rhadinoviruses only, and KSHV only, as indicated. The basic design of this map was patterned after Fig. 1 in reference 28 and is updated from Fig. 1 in reference 26.