FIG 10.
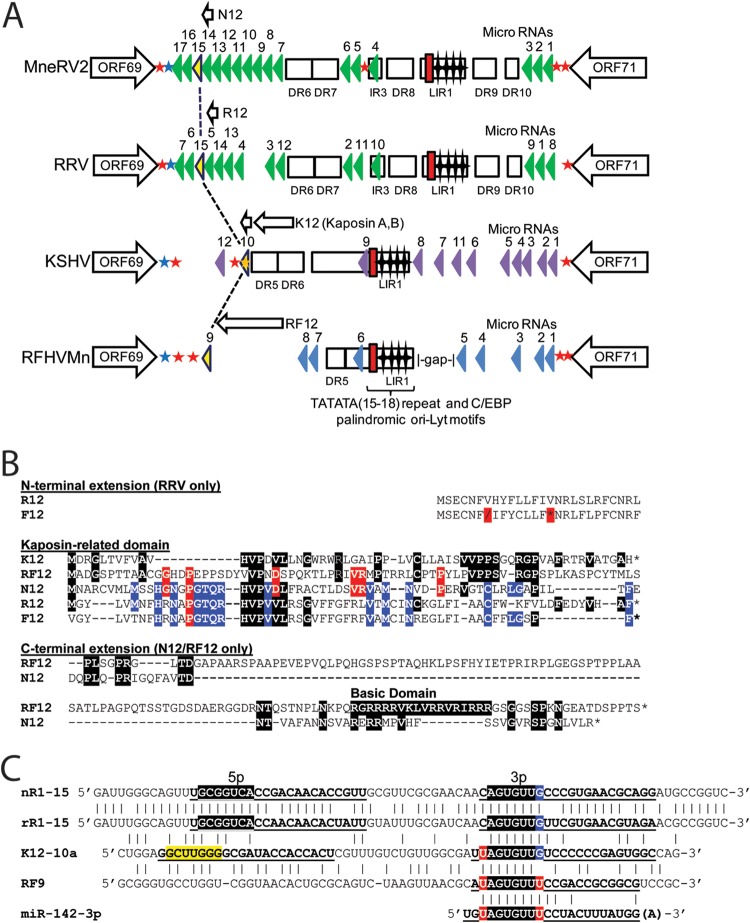
The latency regions of MneRV2 and RRV are highly homologous and encode protein and miRNAs conserved with KSHV and RFHVMn. (A) Schematic representation of the genomic regions of MneRV2, RRV, RFHVMn, and KSHV between ORFs 69 and 71; the presence of encoded proteins and miRNAs is shown. The positions of the TATATA motif (red box) and C/EBP palindromic motifs (stars) within the long inverted repeat (LIR) regions are indicated (Fig. 8). The known KSHV (purple) and RRV (green) miRNA genes are indicated, as are the predicted MneRV2 (green) and RFHVMn (blue) candidate miRNAs. Homologous miRNAs with identical seed sequences conserved across the four viruses are colored yellow (see panel C). Direct repeats (DR) and putative protein coding ORFs are shown. Poly(A) signals “AAUAAA” (blue star, left to right transcription; red star, right to left transcription) and “AAAUCA” (orange star, right to left transcription) identified in KSHV (42) are shown with homologous signals in the other viruses. (B) Alignment of KSHV ORFK12 Kaposin A (YP_001129428) and positional homologs MneRV2 ORFN12, RRV ORFR12 (AF210726 or, without N-terminal extension, NP_598364), and RFHVMn ORFRF12 (AGY30757). The translated sequence of Fu12 from the MfuRV2 genome sequence is shown but lacks a methionine initiator and contains a shift (\) and stop codon (*) in the reading frame. Residues conserved with K12 are highlighted black; residues conserved with RF12 are highlighted red; residues conserved with N12 are highlighted blue. (C) Alignment of the kshv-miR-K12-10a, rfhvmn-miRc-RF9, mnerv2-miRc-N12-15, and rrv-miR-rR1-15 viral pre-miRNAs with the human cellular miR-142-3p. The 5p and 3p mature miRNAs are indicated (bold, underlined). The conserved seed sequences are highlighted in black with 5′ and 3′ residues highlighted in red or blue that would constitute part of the seed sequence depending on processing. The nonconserved K12-10-5p seed is highlighted in yellow.