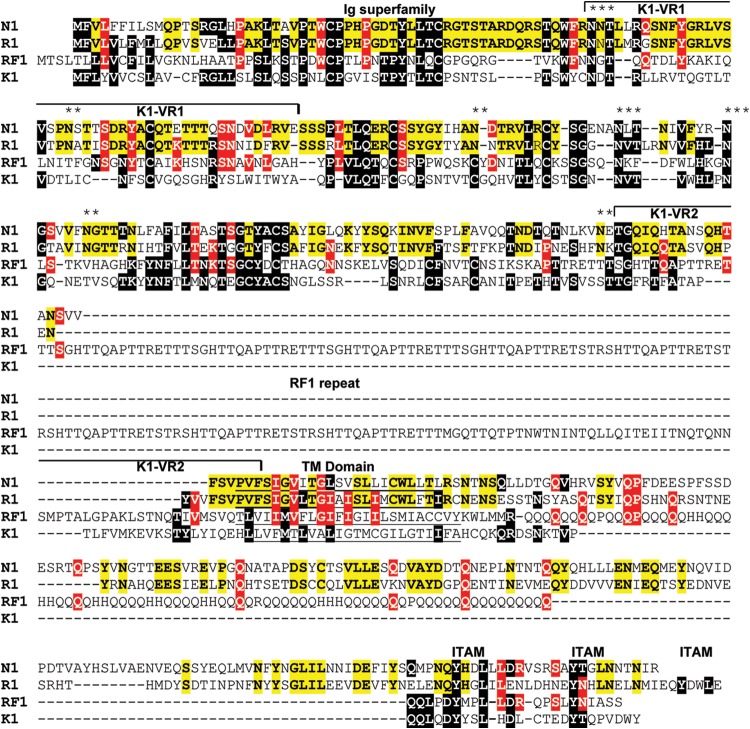
FIG 2.
Alignment of the left-end K1 homologs. The leftmost ORFs in the genomes of MneRV2, RRV26-95, RFHVMn, and KSHV, corresponding to ORFs N1, R1, RF1, and K1, respectively, were aligned. The RRV17577 R1 sequence varies from the RRV26-95 sequence by only 2 aa. K1 residues conserved in other sequences are highlighted in black. Residues conserved between RV1 and RV2 rhadinovirus homologs are highlighted in red, and residues conserved between the macaque RV2 viruses MneRV2 and RRV26-95 are highlighted in yellow. The N-terminal domain related to the immunoglobulin receptor and the immunoglobulin receptor tyrosine-based activation motifs (ITAM) (61) are indicated, and the variable regions (VR1 and VR2) identified in KSHV subtypes (62) are shown. Residues predicted to form a hydrophobic transmembrane (TM) domain and the position of the repeat domain within the RF1 sequence are indicated. Conserved potential N-linked glycosylation sites are shown with asterisks (**, conserved between MneRV2 and RRV; ***, conserved between RV1 and RV2 rhadinoviruses).