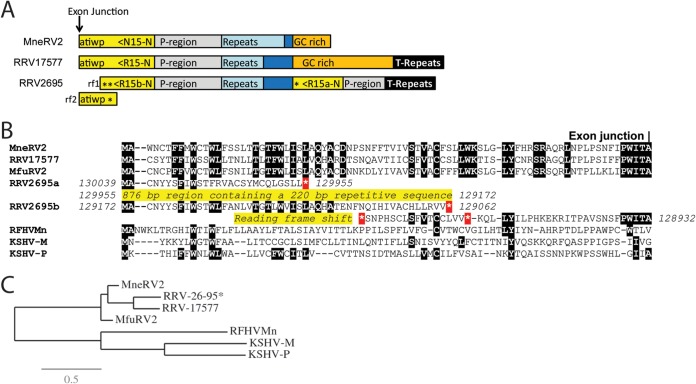
FIG 3.
Comparison of the right-end K15 homologs. (A) Graphical representation of the structure at the right end of the MneRV2, RRV26-95, and RRV17577 genomes with the N-terminal exon of the K15 homolog shown (yellow), with the direction of translation and the amino acid sequence immediately upstream of the exon junction. The presumed promoter region (P-region; gray), repeat region (light blue), GC-rich region (orange), and terminal repeat (T-repeat) region (black) are indicated. The duplicated N-terminal domains of RRV26-95 R15 are shown (asterisk indicates a stop codon). Reading frames 1 and 2 (rf1 and rf2) of the N-terminal domain of RRV26-95 R15b are shown, indicating the reading frameshift disrupting the R15b sequence. (B) Alignment of the N-terminal sequences of the K15 homologs of KSHV (M and P variants), RFHVMn, RRV17577, RRV26-95, and MfuRV2. The conserved exon junction is indicated. The duplicated N-terminal domains of RRV26-95 R15 are shown with the disrupted reading frames and stop codons (*). (C) Maximum likelihood analysis of the complete amino acid sequences of the fully spliced K15 homologs: KSHV-M (AAD45296), KSHV-P (AAK72632), RFHVMn (AGY30764), RRV17577 (40), MfuRV2 (AY528864), RRV26-95 (AF210726; lacks the disrupted N-terminal exon), and MneRV2. The numbers of substitutions per site are indicated.