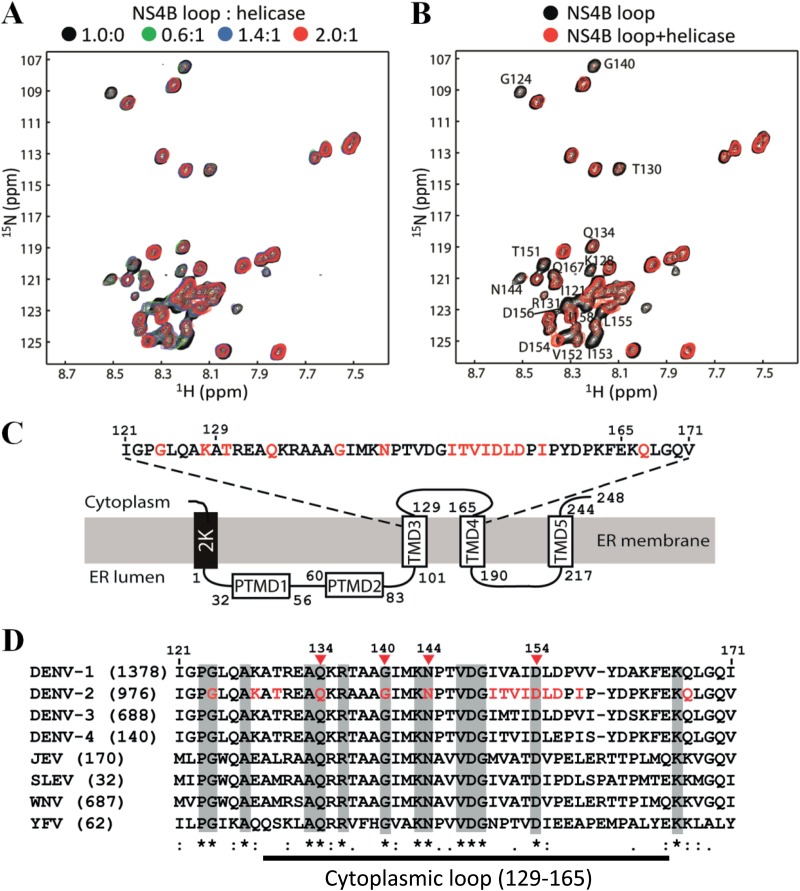
FIG 6.
Chemical shift perturbation analysis of the NS3-NS3B interaction. The recombinant NS3 helicase domain (Fig. 2A) and NS4B (residues 121 to 171) (Fig. 5A) were used to perform chemical shift perturbation. (A) Overlay of the 1H-15N-HSQC spectra of the NS4B cytoplasmic loop in the absence and presence of different amounts of helicase. A uniformly 13C/15N-labeled NS4B loop at a 0.2 mM concentration was used in the study. All the experiments were conducted at 298 K. (B) Residues affected by helicase binding. Overlay of 2D 1H-15N-HSQC of labeled NS4B (residues 121 to 171) in the absence (black) and presence (red) of helicase/NS4B loop at a molar ratio of 1:2 is shown. The affected residues are shown with residue name and number. (C) Schematic diagram of the topology of NS4B from DENV2. Residues identified from the chemical shift perturbation experiment are indicated in red. (D) Sequence alignment of the NS4B fragment (residues 121 to 171) among different flaviviruses. The numbers in parentheses are the numbers of NS4B sequences that were used for the alignment. Sequences were downloaded from the National Center for Biotechnology Information (NCBI) protein database. The alignment was performed using CLC Main Workbench software (CLC bio). The amino acid positions of NS4B are numbered according to the DENV2 NGC strain (GenBank number AF038403). The flavivirus-conserved residues are shaded in gray. The amino acids identified from the chemical shift perturbation analysis are shown in red in the DENV2 sequence, among which four residues (indicated by arrowheads), belonging to the cytoplasmic loop (defined as residues 129 to 165, according to Miller et al. [18]), are strictly conserved across flaviviruses.