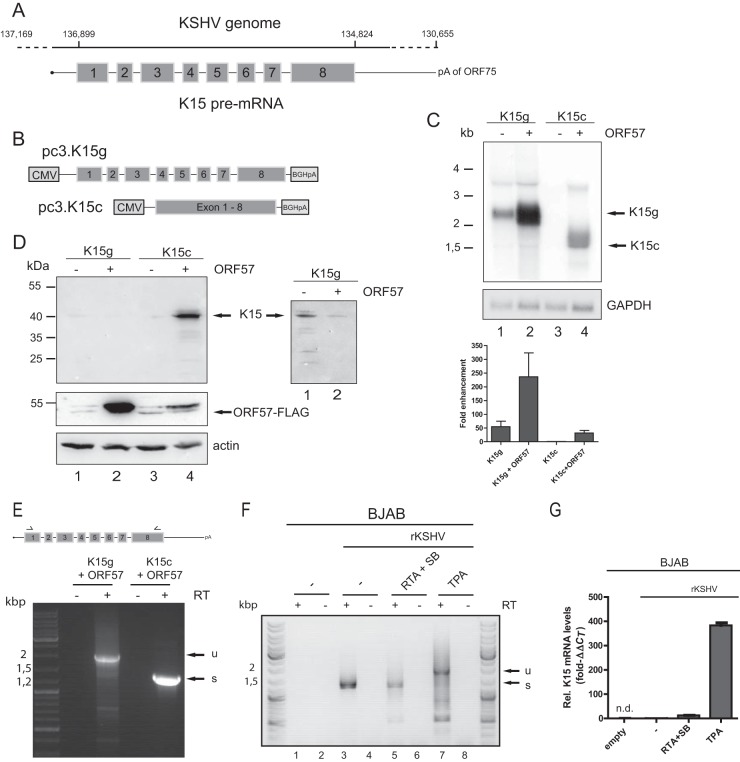
FIG 1.
ORF57 promotes K15 expression at the RNA and protein level. HEK293T cells were cotransfected with pc3.K15g or pc3.K15c and ORF57 cDNA. Total RNA and protein were harvested 40 h posttransfection and analyzed by Northern and Western blotting. Transfection efficiency was monitored by cotransfection of an eGFP-encoding plasmid and measured by flow cytometry in all experiments (data not shown). (A) Depiction of the K15 gene in the viral genome. The coordinates in the genome are given according to NCBI accession number AF148805.2. The outer left coordinate represents the start of the terminal repeat. The dashed line indicates the uncharacterized transcriptional start site. K15 uses the poly(A) site of ORF75. The K15 pre-mRNA is drawn with exons (gray boxes; in scale) and introns (not in scale). (B) Scheme of the pcDNA3.1 K15 expression plasmids. For further details, see the text. (C) Northern blot analysis of total RNA from the indicated cotransfection experiments. The 32P-labeled probe is directed against K15 exon 8. As a loading control, the blot was rehybridized with a GAPDH specific probe. On the left, a size marker in kilobases is indicated, and the RNA species are marked on the right. The Northern blots were quantified by phosphorimager analysis. The K15 signal was normalized to the GAPDH loading control. The expression of K15c without ORF57 was set to 1. Quantifications and standard deviations represent three independent experiments. (D) Western blot with protein extracts from the indicated cotransfections. Detection was performed with a K15 antibody raised against epitopes encoded in exon 8. Actin served as a loading control. The lysates were also used to detect ORF57-FLAG with an anti-FLAG antibody. A size marker in kilodaltons is indicated on the left, and the K15- and ORF57-specific band is indicated on the right. The inset on the right shows weak K15 expression after longer exposure times from the genomic constructs. (E) RNA from the indicated cotransfections was analyzed by RT-PCR. The primers bind in K15 exon 1 and exon 8, illustrated by arrows. The addition of the RT enzyme is depicted. A DNA size marker is shown on the left. u, unspliced; s, completely spliced. (F) BJAB cells harboring a recombinant KSHV genome were reactivated as indicated. The RNA was analyzed as in panel E. (G) The RNA from panel F was subjected to RT-qPCR by using a K15-specific probe. The value for nonreactivated BJAB rKSHV was set to 1. n.d., not detected.