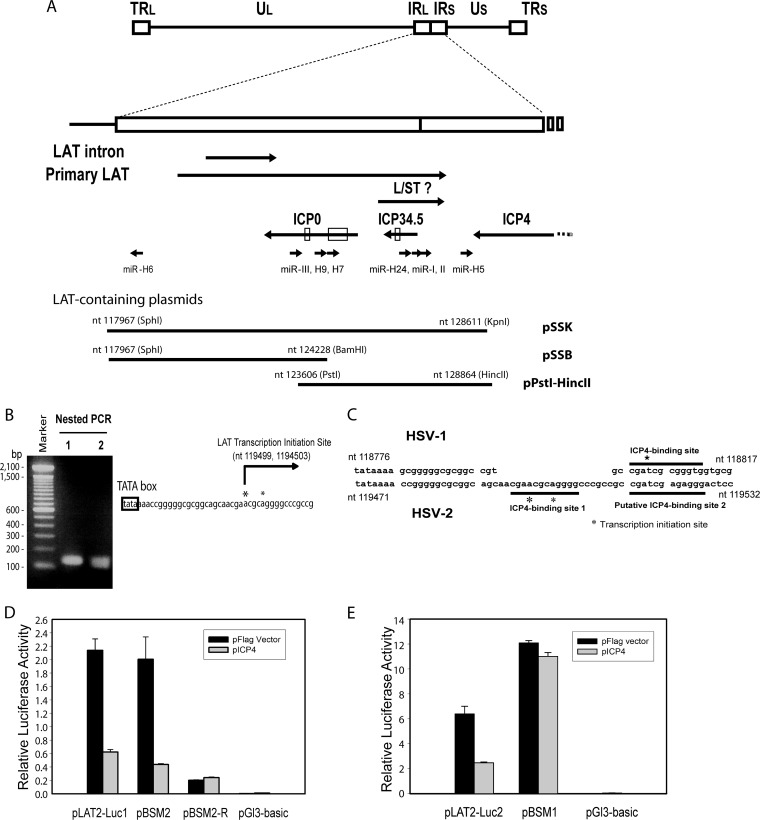
FIG 1.
Identification of HSV-2 LAT ICP4-binding sequence. (A) Schematic diagram of HSV-2 LAT region. The HSV-2 L/ST homolog is labeled. The relative positions of the inserts in LAT-containing plasmids, including pSSK, pSSB, and pPstI-HincII, are shown. (B) Mapping of HSV-2 LAT transcription initiation site. The HSV-2 LAT transcription start sites were mapped to nt 119499 (which was more dominant) and nt 1194503 with a 5′ RACE kit and total RNAs from HSV-2-infected Vero cells. (C) Comparison of the HSV-2 and HSV-1 LAT transcription initiation regions. Two HSV-1 ICP4-binding motifs, including putative ICP4-binding site 1 and putative ICP4-binding site 2 (underlined), were identified near the LAT transcription initiation site. (D) Mutation of putative ICP4-binding site 2 does not affect HSV-2 ICP4-mediated repression of LAT promoter reporter activity in cells transfected with LAT promoter reporters with or without pICP4, indicating that putative ICP4-binding site 2 does not function to limit LAT expression. pLAT2-Luc1 contains LAT sequences ranging from bp −392 to bp +239 relative to the dominant LAT transcription initiation site (nt 119499). pBSM2 contains LAT sequences ranging from bp −392 to bp +239 relative to the dominant LAT transcription initiation site (nt 119499), with substitution mutations in putative ICP4-binding site 2. pBSM2-R contains the same insert as pBSM2, oriented in the reverse direction, in the pGL3-Basic reporter. Firefly luciferase activity was normalized with cotransfected pRL-Ts, a Renilla luciferase reporter. (E) Mutation of putative ICP4-binding site 1, which overlaps the LAT transcription start sites, abolishes inhibition of the LAT promoter reporter activity by HSV-2 ICP4. LAT promoter reporters were cotransfected with or without pICP4 in 293 cells. pLAT2-Luc2 contains LAT promoter and putative ICP4 binding site 1 sequences ranging from bp −392 to bp +8 relative to the dominant LAT transcription initiation site (nt 119499). pBSM1 contains LAT sequences ranging from bp −392 to bp +8 relative to the dominant LAT transcription initiation site (nt 119499), with substitution mutations at putative ICP4-binding site 1.