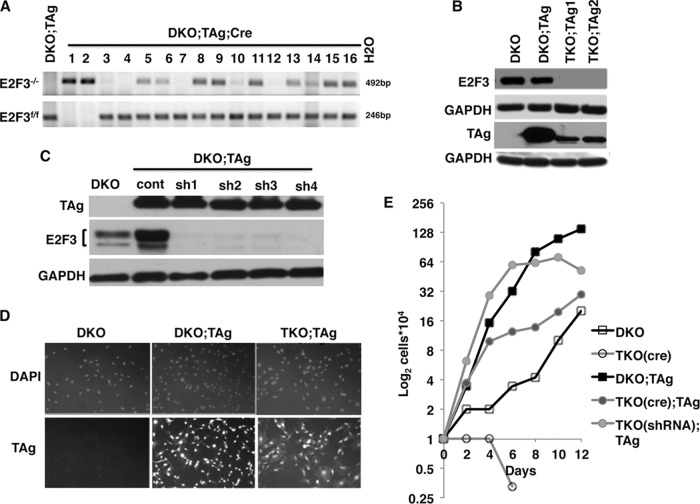
FIG 2.
Generation and characterization of cells lacking E2F3 in the presence of TAg. (A) PCR-genotyping analysis to determine the presence or absence of E2F3 in clones of cre-treated DKO;TAg MEFs. DNA from cells containing a successfully deleted E2F3 allele (E2F3−/−) produced an amplified fragment of 492 bp, while those where the floxed allele (E2F3f/f) remained intact produced a 246-bp fragment. DNA from cells where only one copy of E2F3 was removed produced both fragments upon PCR amplification. (B and C) Western blot analysis confirms the absence of E2F3 and presence of TAg protein in the TKO;TAg clones obtained by cre-recombinase (B) or in pools of MEFs expressing E2F3-specific shRNAs (C). Four different E2F3-specific shRNAs (sh1, sh2, sh3, and sh4) were used with a nontargeting control shRNA (cont). GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (D) Analysis of TAg expression and localization by immunofluorescence using hamster anti-TAg primary antibody and a fluorophore-attached anti-hamster secondary antibody. DKO MEFs not expressing TAg are shown as negative controls. Negative controls without primary antibody incubation produced no significant signal and are not shown. (E) Representative examples of cell growth curves upon depletion of E2F3 from DKO or DKO;TAg MEFs via the use of either cre-recombinase or RNAi. The data shown represent averages of the results of two separate experiments performed in duplicate. TKO(cre);TAg data represent the averages of the results from the two clones obtained via cre-recombinase, and TKO(shRNA);TAg data represents the averages of the results from two pools of cells generated by two different E2F3-specific shRNAs.