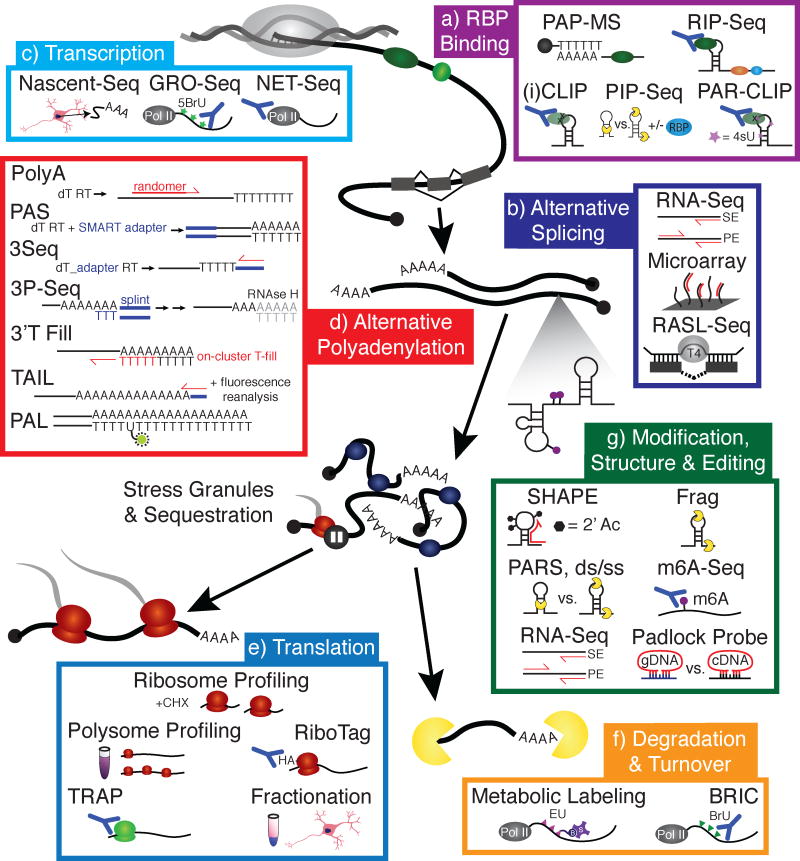
Figure 1. High-throughput sequencing enables quantification of RNA processing steps on a global scale.
a | RBP Binding. Poly(A) purification and liquid chromatography mass spectrometry (PAP-MS) is a method that involves poly(A) selection of RBP-bound RNA followed by proteomic analysis to identify mRNA-bound proteins, enabling the identification of novel RBPs. RNA immunoprecipitation (RIP) is a method for identifying whole transcripts associated with an RBP by immunoprecipitating the RBP with bound RNA, then subjecting the isolated RNA to RNA-seq analysis. Cross-linking immunoprecipitation (CLIP)-seq and individual nucleotide resolution CLIP (iCLIP) improve on the resolution of RIP-Seq by utilizing UV to crosslink RNAs to protein. This enables both more stringent washing to reduce false positives, and a digestion step that reveals specific RBP target regions and motifs. Photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) is similar to iCLIP, but crosslinking efficiency is increased with the metabolic labeling of RNA by 4-thio-uracil (4sU). Protein interaction profile (PIP)-Seq characterizes the structural dependence of RBP-RNA interactions through the inclusion of proteinase K dependent libraries using proteinase K.
b | Alternative Splicing. Splicing inclusion and exclusion events can be detected by a simple RNA-Seq experiment, either single end (SE) or paired end (PE) reads, provided sufficient read depth at exon-exon and exon-intron junctions, and does not require the splicing event to be annotated. Microarrays can also be used to detect splicing changes, and are more sensitive to detecting events in lowly expressed transcripts compared to RNA-Seq; however, the assay is limited to the number of events on an array, as well as prior knowledge of an inclusion/exclusion event. A less comprehensive but very sensitive technique, RNA-mediated oligonucleotide Annealing, Selection, and Ligation with sequencing (RASL-seq), utilizes a ligation reaction to detect an event based on ligation of oligomers complementary to alternative exon-exon junctions.
c | Transcription. Nascent-Seq involves the sequencing of nascent RNAs isolated from the nucleus using centrifugation and fractionation. Global run-on sequencing (GRO-Seq) involves pausing of the transcription machinery, then reinitiation with the addition of brominated nucleotides which are incorporated into nascent transcripts and facilitate immunopurification of the nascent RNAs by antibodies specific for the 5-bromouridine (5BrU). To avoid transcription pausing, native elongating transcript sequencing (NET-Seq) employs immunopurification of RNA polymerase II (pol II) with its associated transcripts for sequencing.
d | Alternative Polyadenylation. Poly(A) site usage can be determined by several techniques, the majority of which involve positive selection by oligo(d)Ts and sequencing into the poly(A) tail. PolyA-Seq involves oligo(d)T-primed first strand synthesis and randomer-primed second strand synthesis. Poly(A) site sequencing (PAS-Seq) also uses oligo(d)T-primed first strand synthesis, but with the inclusion of a SMART adapter added at the end of first strand synthesis, reducing the need for internal randomer priming. 3Seq takes a somewhat modified approach, with oligo(d)T-primed first strand synthesis that includes an adapter for second strand synthesis. A problem with internal priming is the risk that the PCR will not proceed all the way to the poly(A) site, making it impossible to determine alternative polyadenylation. Furthermore, oligo(d)T priming risks polymerase slippage. 3P-Seq avoids this with the use of a splint ligation to attach adapters, while 3′T-fill fills in the poly(A) tract with d(T)s on the flow cell of the sequencing instrument prior to initiation of sequencing. In addition to alternative poly(A) site identification, poly(A) length can be critical in the case of RBPs that bind the poly(A) tail. Poly(A) size can be determined by TAIL-Seq, which utilizes special software to reanalyze the fluorescence signal on the flow cell to eliminate false ‘T’ base calls due to residual signal from reading the poly(A) tract. Alternatively, poly(A) tail length sequencing (PAL-Seq) utilizes stoichiometric incorporation of a modified uridine during poly(A) sequencing that can be fluorescently tagged, identifying tail length as proportional to fluorescence intensity.
e | Translation. Methods for identifying actively translating transcripts involve purification of ribosomes or polysomes by fractionation and sequencing of the associated RNAs, as in ribosome and polysome profiling. Ribosome labeling has enhanced these techniques. In a method similar to CLIP, tagged ribosomes can be immunoprecipitated and the associated RNAs sequenced, as in RiboTag. Alternatively, lineage-specific promoters driving EGFP-labeled ribosomes allow for cell-specific isolation of polysomes, as in TRAP-Seq, further enhancing the resolution of nascent RNA sequencing. One aspect of translational control is localization of the translation machinery and associated RNAs, which can be probed by compartmental fractionation and either sequencing or proteomic analysis of each fraction.
f | Degradation & Turnover. Measuring rates of global mRNA synthesis and degradation is most commonly carried out with a pulse-chase experiment to label nascent transcripts followed by the observed loss of label as transcripts are degraded. Metabolic labeling can be performed with 5-ethynyl uridine (EU), which can be biotinylated with click chemistry for purification of pulse-labeled transcripts, or with 4-thiouridine which can also be biotinylated. Similarly, RNA can be pulsed with 5-bromo-uridine (BrU) in BrU immunoprecipitation chase-deep sequencing analysis (BRIC-Seq). These techniques require an immense amount of input material, making it a challenging assay for difficult-to-obtain cell types, such as iPS-derived neurons.
g | Modifications, Structure & Editing. High-throughput methods for determining secondary structure include selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE)-Seq, which involves the acetylation of specific bases in single-stranded loops and bulges, blocking cDNA synthesis and enabling single nucleotide resolution of certain secondary structures. To glean information on regions of secondary structure, differential digestion with single and double-strand specific nucleases can be utilized. Fragmentation sequencing (Frag-Seq) utilizes a single-strand-specific nuclease to identify regions of RNA involved in base pairing structures. Parallel analysis of RNA structure (PARS)-Seq compares sequencing libraries of single strand (ss) and double strand (ds)-specific nuclease-treated RNA to identify secondary structure. To identify RNA editing events, such as A-to-I editing, RNA-Seq often suffices. However, to probe a specific site, padlock probes can be designed to compare the identity of a single base between cDNA generated from RNA to the genomic DNA. m6A-Seq enables low-resolution sequencing of m6A modifications through nucleobase IP.