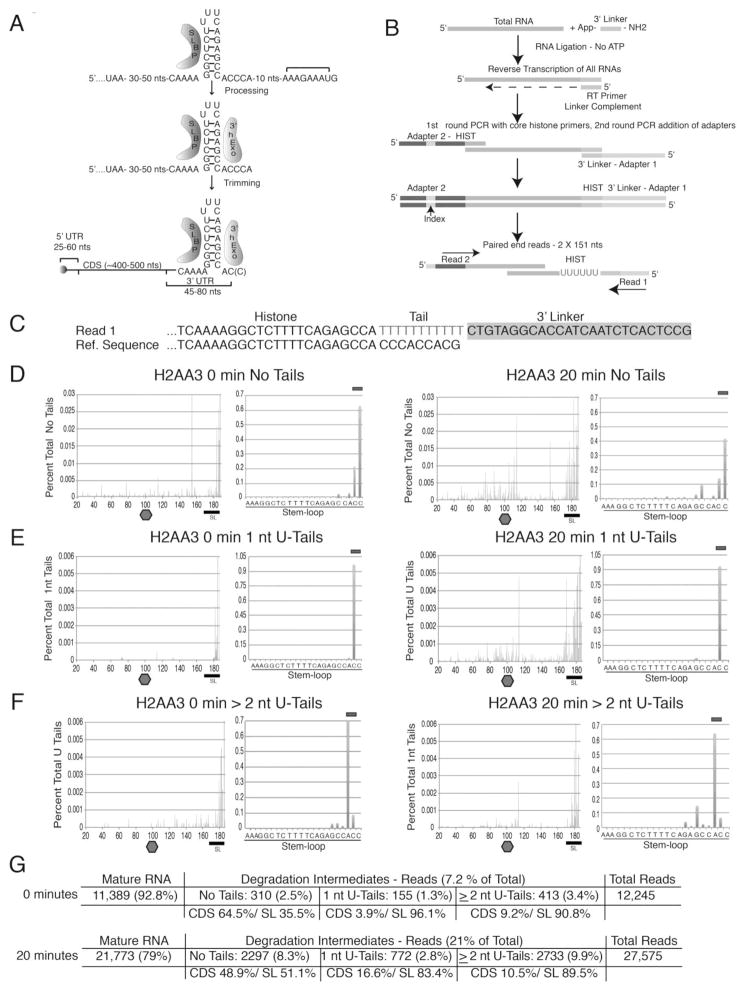
Figure 1. Strategy to detect histone mRNA degradation intermediates.
(A) Histone pre-mRNA processing steps resulting in the mature transcript bound by SLBP and 3′hExo.
(B) Diagram of the high-throughput sequencing strategy for 3′-end analysis of histone mRNA degradation intermediates (see the Supplemental Experimental Procedures). RNA ligated to a preadenylated linker is reverse transcribed with the linker complement (LC), followed by limited PCR with a histone-specfiic primer. A second round of limited PCR adds Illumina specific adaptors.
(C) Alignment of a sequence of a U-tailed histone mRNA ligated to the linker (top) with a histone reference sequence (bottom).
(D–F) The distribution of the 3′ ends of reads from the HIST2H2AA3 mRNA before (left) or after (right) 20 min HU treatment. The 3′ ends of the untailed RNAs (D), RNAs with 1-nt U-tail (E), and RNAs with U tails of ≥ 2 nt (F) were plotted. The stop codon was set at position 100, and the top bar indicates the mature mRNAs ending in AC or ACC (D), ACU (E) or AUU or ACUU (F) after the stem. On the right are the tails in the SL on an expanded scale.
(G) Table summarizing the number of mature and tailed RNAs.