Figure 4.
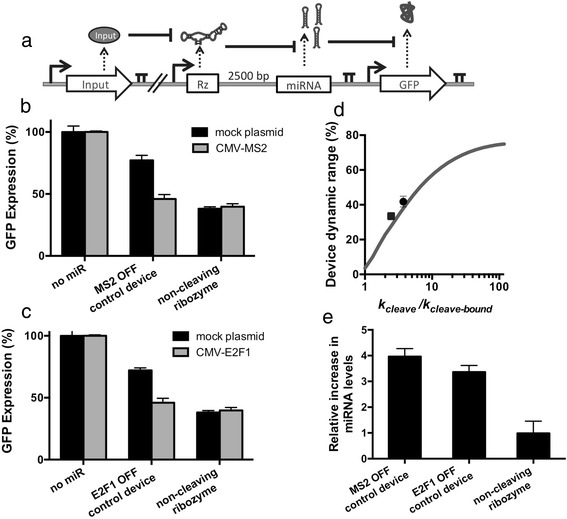
Protein-responsive RNAi-based OFF control devices. (a) Schematic overview of the genetic circuit used to characterize protein-responsive OFF control devices. The OFF control devices and GFP were stably integrated and expressed from identical expression cassettes as described in Figure 2a, except that the theophylline-responsive ribozyme was replaced with an MS2- or E2F1-responsive ribozyme switch. A plasmid encoding the expression of DsRed only (mock plasmid) or the protein ligand (MS2 or E2F1) was transfected into the cell lines to measure activity of the OFF control device. (b-c) GFP expression levels under the control of the MS2-responsive (b) or E2F1-responsive (c) OFF control device. No miR and miRNA only represent constructs with a protein-responsive ribozyme switch and no downstream miRNAs (negative control) and a construct with a non-cleaving ribozyme upstream of a GFP-targeting miRNA (positive control), respectively. Error bars represent ±1 standard deviation of at least three independent experiments. (d) Device dynamic ranges for the OFF control devices as a function of the ratio of cleavage rates (k cleave/k cleave-bound). Grey line: model; circle: theophylline-responsive device; square: MS2-responsive device. Device dynamic range is reported as the percent change in GFP expression in the absence and presence of theophylline. (e) Relative increase in miRNA levels after addition of protein ligand. Relative miRNA levels are reported normalized to the miRNA levels measured from identical cell lines in the absence of protein ligand. Error bars represent ±1 standard deviation of three independent experiments.
