Abstract
Garden asparagus (Asparagus officinalis L.) is a commercially important crop species utilized for its excellent source of vitamins, minerals and dietary fiber. However, after harvest the tissue hardens and its quality rapidly deteriorates because spear cell walls become rigidified due to lignification and substantial increases in heteroxylan content. This latter observation prompted us to investigate the in vitro xylan xylosyltransferase (XylT) activity in asparagus. The current model system for studying heteroxylan biosynthesis, Arabidopsis, whilst a powerful genetic system, displays relatively low xylan XylT activity in in vitro microsomal preparations compared with garden asparagus therefore hampering our ability to study the molecular mechanism(s) of heteroxylan assembly. Here, we analyzed physiological and biochemical changes of garden asparagus spears stored at 4 °C after harvest and detected a high level of xylan XylT activity that accounts for this increased heteroxylan. The xylan XylT catalytic activity is at least thirteen-fold higher than that reported for previously published species, including Arabidopsis and grasses. A biochemical assay was optimized and up to seven successive Xyl residues were incorporated to extend the xylotetraose (Xyl4) acceptor backbone. To further elucidate the xylan biosynthesis mechanism, we used RNA-seq to generate an Asparagus reference transcriptome and identified five putative xylan biosynthetic genes (AoIRX9, AoIRX9-L, AoIRX10, AoIRX14_A, AoIRX14_B) with AoIRX9 having an expression profile that is distinct from the other genes. We propose that Asparagus provides an ideal biochemical system to investigate the biochemical aspects of heteroxylan biosynthesis and also offers the additional benefit of being able to study the lignification process during plant stem maturation.
Introduction
Heteroxylans represent a major family of non-cellulosic polysaccharides in dicot secondary walls and monocot primary walls [1]. They are composed of a linear backbone of 1,4-linked β-D-xylose (Xyl) residues substituted with variable side branches that are mostly composed of α-D-glucuronic acid (GlcA), 4-O-methyl-α-D-glucuronic acid (MeGlcA) and/or α-L-arabinofuranose (Araf) depending on the species and tissue types [2]. Thus, heteroxylans are classified according to their backbone substitution patterns as either (methyl)glucuronoxylans ((Me)GXs), glucurono(arabino)xylans (GAXs) or arabinoxylans (AXs). (Me)GXs predominate in the secondary walls of dicot wood and non-commelinoid monocots, the grouping that includes garden asparagus (Asparagus officinalis L.) and many other economically valuable species. However, in the commelinoid monocots, represented by the commercially important cereals (grasses) such as rice, maize, wheat and barley, GAXs are the most abundant non-cellulosic matrix polysaccharides [3]. AXs are particularly rich in cereal endosperm walls, constituting up to 70% (w/w) of the endosperm cell wall in wheat [4].
Heteroxylans are synthesized in the Golgi apparatus (GA) and then transported via post-Golgi secretory vesicles to the plasma membrane where they are deposited into the wall [5]. The biosynthesis requires multiple type II glycosyltransferases (GTs) including xylosyltransferases (XylTs) responsible for generating the β-(1,4)-Xyl backbone and glucuronosyltransferases (GlcATs) and arabinosyltransferases (AraTs) responsible for incorporation of the major side chain residues [6]. In Arabidopsis, several GTs involved in heteroxylan biosynthesis have been identified via mutant analysis. These include members of the CAZy GT family 47 (IRX10 and FRA8/IRX7) [7–9], GT43 (IRX9 and IRX14) [10,11] and GT8 (IRX8, PARVUS and GUX) [12–14]. Among these GTs, IRX9, IRX10, and IRX 14 as well as their homologs (IRX9-Like (L), IRX10-L and IRX14-L) are believed to be essential for the elongation of the xylan backbone [8,10–12,15–17]. Additionally, several gene families responsible for the glycosyl residue substitution by non-glycosyl residues have been reported and include DUF579 for transfer of a methyl group to GlcA on the xylan backbone [18], BAHD involved in ferulic and p-coumaric acid addition [19] and TBL29 catalyzing Xyl acetylation [20,21].
In parallel, in vitro xylan biochemical assays have been developed in order to understand the molecular mechanism of their assembly. Feingold [22] first demonstrated that Asparagus microsomes could transfer D-Xyl to a β-(1,4)-linked xylo-oligosaccharide acceptor but only a single Xyl was reported to be incorporated. Subsequently, processive xylan-XylT activity has been confirmed in various species including maize [23], sycamore and poplar [24], pea [25], Zinnia [26], wheat [27] and Arabidopsis [10,15]. Arabidopsis GT43 mutants (irx9 and irx14) have reduced xylan and xylan XylT activity, providing evidence that they play a role in xylan biosynthesis [15]. Tobacco microsomes from suspension-cultured cells co-expressing both IRX9 and IRX14, but not IRX9 and IRX14 seperately, have been shown to have XylT activity, indicating that these two proteins act cooperatively and may be part of an active xylan biosynthetic complex [28]. A partially purified xylan biosynthetic complex was isolated from wheat seedlings and found to contain GT43, GT47 and GT75 proteins, suggesting their involvement in GAX biosynthesis [29]. Recently, two groups have independently demonstrated that heterologously expressed IRX10 has processive xylan XylT activity. Urbanowicz et al [21] expressed the Arabidopsis IRX10-L in mammalian HEK293 cells and Jensen et al [30] expressed separately a Plantago ovata and the moss (Physcomitrella patens) IRX10 gene in Pichia pastoris and found that the heterologously expressed IRX10 proteins could add multiple Xyl residues onto a xylo-oligosaccharide acceptor. The P. patens homolog was described as displaying “robust” xylan XylT activity with the Arabidopsis and P. ovata homologs showing “lower but reproducible” activity [30]. However, it remains unclear whether the heteroxylan biosynthetic mechanism(s) between dicots and grasses are the same since dicot (and gymnosperm) xylans possess a distinct ‘primer’ oligosaccharide at the reducing end [31] unlike grass xylans that lack this specific sequence [32,33].
The monocot order Asparagales includes many economically important species, some with edible plant parts such as onion, garlic, asparagus and vanilla as well as many cut flower species including orchids, daffodils and irises. The ‘developmentally immature, rapidly growing’ subterraneous shoot (also referred to as the spear) of garden asparagus (Asparagus officinalis L.) is an important agricultural crop consumed for both its dietary fibre values and distinctive flavor. Post-harvest Asparagus ‘quality’, including its texture and flavor, deteriorates relatively quickly and this has been attributed to tissue maturation-related hardening [34]. This hardening is due to secondary wall formation and increased lignification of the secondary walls of the vasculature and supporting structures. This maturation process is also associated with a significant increase in heteroxylans [35] and xylan-pectic polysaccharide complexes [36], suggesting there is active xylan synthesis occurring in the Asparagus spear post-harvest.
In this study, we verified the changes in the polysaccharide composition and lignification process in Asparagus spears. We then biochemically characterized a surprisingly active xylan XylT activity in different stem sections (apical, middle and basal) over post-harvest stages. We found that the highest XylT activity is in the basal section of Asparagus spears, a region that also has high heteroxylan and lignin content. In addition, we generated a reference transcriptome to examine the expression of Asparagus XylT genes throughout the sections and storage stages. Together our results show that Asparagus is an ideal non-commelinoid monocot model system to study heteroxylan biosynthesis, particularly as it offers a tractable system to purify the in vivo protein complex responsible for the biochemical activity that will be essential in elucidating our understanding of the molecular mechanism of xylan synthesis.
Materials and Methods
Plant material
Green garden asparagus (Asparagus officinalis L.) spears were harvested from a local producer (Vizzarri Farm, KooWee Rup, Victoria,Australia) and transported on ice to the laboratory within 2 hr post-harvest. After gentle washing with distilled water, dry spears with a mean butt (bottom of the spear) diameter of 1.0–1.5 cm were cut to approximately 21 cm in length and marked at 7-cm intervals. The spears were subsequently sorted into bunches weighing approximately 600 g (~15 spears per bunch). Each bunch was placed into plastic containers and stored at 4±1°C for up to 16 days with ~95% relative humidity in the dark. Asparagus spears at harvest (0 day) and after 4, 8, 12 and 16 days of storage were cut into three 7-cm sections (apical, middle and basal) and the FW measured. For RNA extraction, the spears were frozen in liquid nitrogen and stored at -80°C prior to processing.
Fresh weight (FW) loss
Weight loss was recorded periodically by weighting 15 spears during storage using a digital 3 decimal place balance (Mettler-Toledo, Melbourne, Australia). The results were expressed as the percentage loss of initial FW.
Visual evaluation of Asparagus spear appearance
The general appearance of the spears was independently assessed by at least 10 trained panelists and scored using a 5 to 1 scale, where 5 = excellent, 4 = very good, 3 = good, 2 = fairly good, 1 = bad. The appearance index was calculated as Σ = appearance scale × percentage of corresponding spears within each class. A minimum value of 3 is required to be considered as commercially acceptable [37].
Determination of spears texture
Asparagus spear texture was determined using a TA-XT2i texture analyzer (Stable Micro Systems, England) fitted with a 5 mm diameter probe. The penetration rate was 1 mm/s with a final penetration depth of 10 mm. The maximum of weight was recorded through the curve. Data were expressed as kg cm-2.
Cell wall polysaccharide and lignin composition analysis
Asparagus spear sections were separately ground with a mortar and pestle in liquid nitrogen and washed successively with 80% ethanol, acetone and methanol to generate alcohol insoluble residues (AIR) as described by Pettolino et al. [38]. The neutral sugar (alditol acetates) and polysaccharide (linkage analysis) compositions of the AIR were determined by GC-MS (Hewlett-Packard HP 6890 GC with a Hewlett Packard 5973 MS, Agilent) following the procedure described by Pettolino et al. [38].
Lignin content was determined gravimetrically as described by Luo et al. [39]. The final lignin-containing residue was weighed and the Klason-lignin content recorded as grams per 100 g FW.
Preparation of microsomal membranes
Microsomes were isolated from Asparagus spears following the procedure of Zeng et al. [40] with minor modifications. Briefly, plant tissues were homogenized with extraction buffer (1 mL/g tissue) containing 50 mM HEPES-KOH (pH 6.8), 0.4 M sucrose, 1 mM dithiothreitol (DTT), 5 mM MnCl2, 5mM MgCl2 and complete EDTA-free proteinase inhibitor cocktail tablet (Roche, Basel, Switzerland) in a kitchen magic bullet blender. The homogenate was filtered through two layers of miracloth (Merck Millipore, Billerica, MA, US) and the filtrate was centrifuged at 3000g for 20 min at 4°C. The supernatant was centrifuged at 100,000g for 30 min at 4°C. The 100,000g pellets were resuspended in homogenization buffer and stored at -80°C. The protein concentration was measured using the BCA protein assay kit (Thermo Scientific, Rockford, IL, US). The bovine serum albumin (BSA) provided in the kit was used as the standard.
Assay of xylan XylT activity by radiolabeling
Xylan XylT activity was measured according to Lee et al. [28]. Microsomes (300 μg of protein) were incubated in a 50 μL reaction mixture containing 50 mM HEPES-KOH (pH 6.8), 0.4 M sucrose, 1 mM DTT, 5 mM MnCl2, 5mM MgCl2 and 1% Triton-100, 0.5 mM Xyl6 and 1.4 μM UDP-[14C]-Xyl (total 15,000cpm) (PerkinElmer, Waltham, CA, US) and 0.25 mM unlabeled UDP-Xyl (CCRC, Athens, Georgia, US). After incubation at RT for 1 hr, the reaction was stopped by adding acetic acid and EGTA (final concentration 0.3 M and 20 mM, respectively). The radiolabeled xylo-oligosaccharides were separated from UDP-[14C]-Xyl by paper chromatography as described by Ishikawa et al. [41]. The terminated reaction mixture was spotted onto a strip of Whatman 3MM paper and developed overnight with the solvent of 95% ethanol: 1 M ammonium acetate (2: 1, v/v) as mobile phase. The radiolabeled Xyl-oligosaccharide products were immobilized on the baseline of the chromatogram and the radioactivity was counted with a Tri-Carb 2100TR liquid scintillation counter (PerkinElmer, Waltham, CA, US) using 4 mL of non-aqueous biodegradable counting scintillant (Amersham Biosciences, Pittsburgh, PA, US). Enzyme assays were performed at least in duplicate.
Anthranilic acid labeling of xylo-oligosaccharides
The assay Xyl and xylo-oligosaccharides (Xyl2-Xyl6, Megazyme, Wicklow, Ireland) were labeled at their reducing termini with anthranilic acid (AA) according to the method described by Alwael et al [42]. Excess derivatisation reagent was removed by mixing with diethyl ether (x3) and the AA-labeled oligosaccharides (Xyl1-6-AA) in the bottom layer were purified using a Sep-Pak C18 cartridge (Waters, Milford, MA, US).
Assay of xylan XylT activity using Xyln-AA as acceptor and RP-HPLC analysis of Xyln-AA products
The assay of XylT activity with Xyln-AA acceptors was performed in a reaction mixture containing 50 mM HEPES-KOH (pH 6.8), 1 mM DTT, 5 mM MnCl2 and 1% Triton-100, 0.25 mM cold UDP-Xyl, 0.1 mM Xyln-AA and 600 μg of microsomal membrane protein in a total volume of 100 μL. After incubation at RT, the reaction was terminated with acetic acid and EGTA (final concentration 0.3 M and 20 mM, respectively) and filtered through a 0.22 μm Ultrafree-MC filter (Merck Millipore, Billerica, MA, US). The products were analyzed by reversed-phase (RP) chromatography on an Agilent 1200 Series HPLC systems and JASCO FP-920 Intelligent Fluorescence Detector (Ex320 nm, Em420 nm). The Xyln-AA products were separated on 2.1 mm × 100 mm, 1.8 μm ZORBAX Eclipse XDB-C18 RP column at a flow rate of 0.5 mL min-1 and a column temperature of 22°C using a gradient program set to: 0 to 2 min (8% B), 2.1 to 20 min (8% to 20% gradient B). Mobile phase A was 50 mM sodium acetate buffer (pH 4.3) and mobile phase B was acetonitrile.
Gel filtration of xylan XylT assay products
The products of the radiolabelling XylT activity (as described above) were mixed with 0.5 ml 50% (v/v) Dowex 1x8 chloride form in water (Sigma-Aldrich, St. Louis, MO, US) and filtered through a 0.8 mL Pierce Centrifuge Tube (Thermo Scientific, Rockford, IL, US). The eluent was fractionated on a Bio-Gel P2 column (Bio-Rad, Hercules, CA, US) and fractions (1.8 ml) were collected. The radioactivity of each fraction (600 μl) was counted with a Tri-Carb 2100TR liquid scintillation counter (PerkinElmer, Waltham, CA, US) 4 mL of biodegradable aqueous scintillant (Amersham Biosciences, Pittsburgh, PA, US).
Treatment of xylo-oligosaccharides with endo-xylanase and β-xylosidase
The XylT assay using the Xyln-AA acceptors was terminated by heating at 100°C for 3 min. The products were then centrifuged at maximum speed in a microfuge for 5 min. Aliquots of the supernatant (10 μL) were saved for treatment with 10units of end-β-(1,4)-xylanase and 10 units of β-xylosidase by mixing with 30 μL 50 mM sodium acetate (pH 5.5,) containing either endo-β-(1,4)-xylanase (Trichoderma viride, GH11(M1), Megazyme, Wicklow, Ireland) or β-xylosidase (Sigma-Aldrich, St. Louis, MO, US), and incubated for 6 hr at 37°C. Digestions were terminated with 0.3 M acetic acid and subjected to RP-HPLC analysis as outlined above.
MALDI-TOF-MS
The AA-labeled xylo-oligosaccharides collected from RP-HPLC were concentrated by SpeedVac and ZipTip (Millipore, Bedford, MA, US). The samples were analyzed on a 7T SolariX hybrid ESI/MALDI-FT-ICR-MS (Bruker, Billerica, Massachusetts, US). Aqueous sample (0.5 μl) was mixed (1: 1, v/v) with the MALDI matrix (saturated 2, 5-dihydroxbenzoic acid (DHB) in 50% acetonitrile) and dried on the stainless steel target plate. The MS was operated in the positive mode using the Broadband acquisition mode, averaging 8 spectra across the mass range = 247–1700 m/z, TD for acquisition was set to 1 M, source accumulation set to 0.001 s, ion accumulation set to 0.4 s and ion cooling set to 0.05 s. The laser spot size was set to large and laser power set at 30%, a total of 1000 laser shots were fired at a rate of 2 kHz per spectrum.
RNA-Seq analysis of Asparagus spears and identification of candidate xylan synthesis genes
Total RNA from fresh and 4 day-old cold-stored Asparagus spears was extracted using the Isolate Plant RNA Mini Kit (Bioline, London, UK) and treated with DNase I (Invitrogen, Carlsbad, California, US) according to the manufacturer’s instructions to remove genomic DNA contamination. Total RNA samples were then quantified using a Nano-Drop (Thermo Scientific, Rockford, IL, US) and 10 μg total RNA submitted to AGRF (Melbourne, Victoria, Australia) for library construction (without size selection) and sequencing on a HiSeq 2000 (Illumina). Raw RNA-seq data from a single lane (191,179,245 read paired-end reads) was subjected to a quality control step involving adaptor removal and sequence trimming using Nesoni (http://www.vicbioinformatics.com/software.nesoni.shtml, min. length 30, quality 24, first 10 bp trimmed) and then de novo-assembled using Trinity [43]. The assembly was then scaffolded using CAP3 [44] (default settings) for each Trinity component.
A quantitative RNA-Seq experiment was also conducted using four biological replicates of apical, middle and basal section from fresh Asparagus spears, each replicate comprising tissue sections from five separate spears. Samples were run on an Illumina HiSeq 2000 in two lanes in a single flow-cell. All samples were sequenced in both lanes. Reads from each sample from each lane were merged and then filtered for quality and adaptor removal as per the reference sample. Reads for each sample were then quantitated independently against the reference transcriptome using RSEM v1.2.8 [45]. Orphaned reads were not used for quantitation. EBSeq v1.1 [46] was used to identify differentially expressed genes.
Candidate xylan biosynthetic genes and several known secondary cell wall regulation genes were identified in the Asparagus reference transcriptome by BLASTX searches (NCBI BLAST+ v2.2.25) using Arabidopsis sequences as bait.
Sequence alignment and phylogenetic analysis
Sequence alignments and phylogenetic analysis was performed using Clustal X 2.1 [47]. IRX proteins from Arabidopsis thaliana (At), Asparagus officinalis (Ao), rice (Oryza sativa, Os) and moss (Physcomitrella patens, Pp) were aligned using Clustal X 2.1 and used as input to produce a neighbor-joining tree with 1000 bootstrap replicates using default settings.
Semi-quantitative RT-PCR
One microgram of total RNA from each section was reverse-transcribed to cDNA using Tetro cDNA synthase kit (Bioline, London, UK) as per the manufacturer’s instructions. Semi-quantitative PCR was performed using My-Taq red DNA polymerase (Bioline, London, UK) with 1 μl cDNA as template. Thirty cycles of the following amplification program was used: 95°C 15 sec, 55°C 15 sec, 72°C 10 sec. PCR primer sequences are listed in S2 Table.
Results
Changes in fresh weight (FW), visual quality, texture and lignin content of garden asparagus spears after harvest
Green Asparagus spears stored at 4°C post-harvest had a reduction in FW of ~10% after 16 days of storage time (Fig 1A). The visual appearance index also dropped significantly (~50%; Fig 1B), correlating with the loss of FW. No statistically significant difference in wet weight was observed between 12–16 days of storage. However, during this time the appearance index dropped from ~3.5 (commercially acceptable) to ~2.5 (not commercially acceptable). Spear texture remained largely unchanged in the apical and middle sections (Fig 1C). Interestingly, the basal sections of the Asparagus spears had increased texture within the first 8 days of storage which then dropped between 8–16 days of storage. Lignin content of Asparagus spears steadily increased over storage time (Fig 1D) with the highest content in the basal section where the spear diameter is the largest, while the apical segment of the spear had the least lignin. These observations are consistent with previous reports [48].
Fig 1. Physiological characteristics of fresh and stored Asparagus spears during low temperature (4°C) storage.
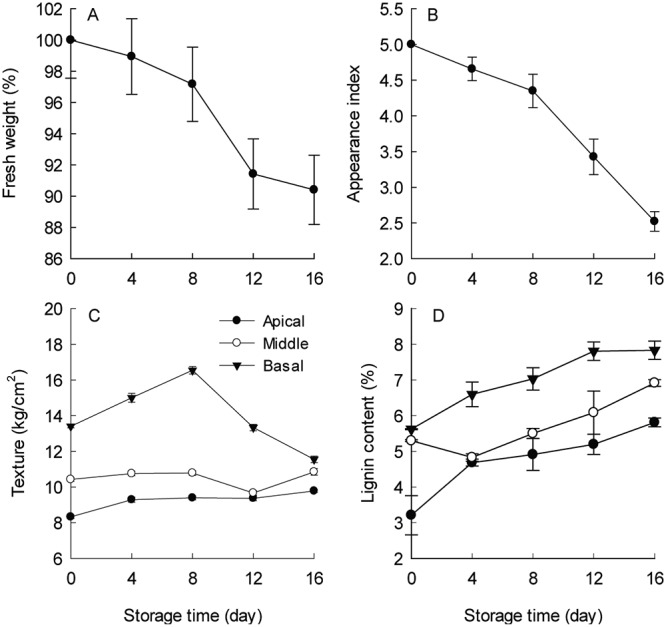
(A) Changes in fresh weight, (B) visual appearance index, (C) texture (firmness) and (D) lignin content. The data are the average of fifteen Asparagus spears.
Changes in cell wall components in garden asparagus spears after harvest
The polysaccharide composition of different spear regions (apical, middle and basal) was determined by linkage analysis (summarized in Table 1, see also S1 Table). The major cell wall components of Asparagus spears include pectic arabinan, heteroxylan, xyloglucan (XG), heteromannan, cellulose and arabinogalactan (AG) (Type I and Type II) (Table 1). Strikingly, the different sections of the spear had different wall polysaccharide compositions. In fresh spears the apical section was composed mainly of arabinan (23%) and only modest amounts of heteroxylan (17%). In contrast, the basal section of the spear contained 55% heteroxylan, coincident with a harder texture and higher lignin content. Furthermore, the substitution levels of xylan, an indicator of solubility, were substantially lower in the basal section and increased from base to apex in both fresh and stored spears (S1 Table). The storage of Asparagus spears at 4°C for 16 days led to a significant decrease in the amount of arabinan, especially in the apical section of the spear where levels fell from 23% in fresh tissue to 18% in the stored tissue (Table 1). In contrast with arabinan, heteroxylan content increased in the apical and middle sections upon storage at 4°C, whereas there was no obvious change in the basal section. Heteromannan levels were similar and highest in the apical/middle sections for both fresh and stored spears. Interestingly, cellulose content in fresh apical and basal sections are lower to that of the middle section but upon storage these levels become similar over all sections.
Table 1. Cell wall neutral polysaccharide components (mol percent) in fresh and stored asparagus spears.
| Fresh | Stored | |||||
|---|---|---|---|---|---|---|
| Bottom | Middle | Top | Bottom | Middle | Top | |
| Arabinan | 4 | 13 | 23 | 4 | 11 | 18 |
| Type I AG | 5 | 10 | 10 | 2 | 3 | 4 |
| Type II AG | 2 | 3 | 3 | 2 | 4 | 5 |
| Glucuronoarabinoxylan | 55 | 23 | 17 | 54 | 32 | 19 |
| Heteromannan | 6 | 13 | 11 | 8 | 15 | 15 |
| Xyloglucan | 5 | 7 | 6 | 3 | 5 | 8 |
| Cellulose | 12 | 20 | 14 | 19 | 17 | 18 |
| Unassigned | 12 | 11 | 16 | 8 | 13 | 13 |
A comparison of xylan XylT activity among different plant species
Arabidopsis is an excellent system to study xylan biosynthesis via mutant analysis, however, biochemical investigation is challenging due to the limited availability of tissues and the inherently low activity of the xylan XylT in in vitro microsomal preparations. The Asparagus spear system was therefore examined for xylan XylT activity by comparison with existing models in dicots (Arabidopsis) and grasses (barley). Thus, a biochemical assay was optimized using microsomal membranes with either radiolabeled (UDP-[14C]-Xyl) or unlabeled UDP-Xyl and xylo-oligosaccharides (either free or the reductively aminated anthranilic acid (AA) derivatives).
Using a paper chromatography assay measuring incorporation of [14C]-Xyl into a Xyl6 acceptor, Arabidopsis and particularly Asparagus microsomes showed XylT activity (Fig 2). In contrast, no activity could be detected in microsomes prepared from barley coleoptiles. Arabidopsis microsomes incorporated ~2% of total radiolabelled Xyl whereas Asparagus incorporated ~26%, i.e. a 13-fold higher amount than that of Arabidopsis.
Fig 2. Comparison of xylan XylT activities in microsomes from Arabidopsis, Asparagus and barley (Hordeum vulgare).
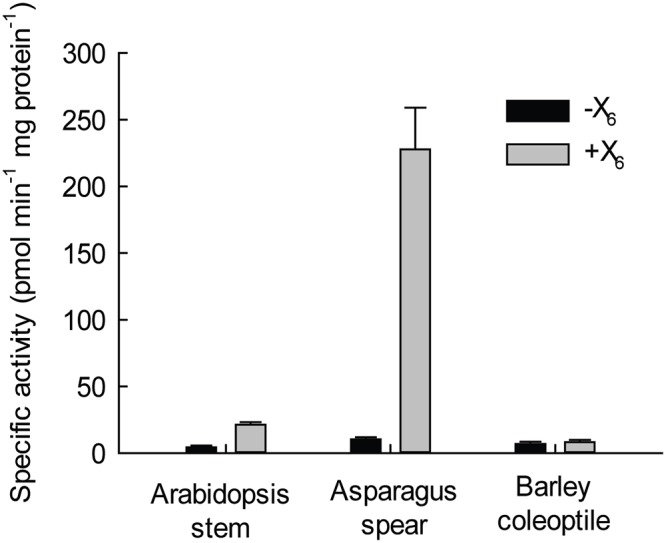
Microsomes from Arabidopsis stem, Asparagus spear and etiolated barley seedlings were isolated and the XylT activities were analyzed in the absence (-; black) and presence (+; grey) of the exogenous acceptor Xyl6 according to the Materials and Methods.
Biochemical properties of Asparagus xylan XylT activity
Asparagus xylan XylT activity was measured under standard radioactive assay conditions to be optimal at ~24°C and around pH 7 and (Fig 3A and 3B, respectively). The activity was time-, protein-, acceptor (Xyl6) and substrate (UDP-Xyl) concentration- (Fig 3C, 3D, 3E and 3F, respectively) dependent. The Michaelis-Menten constant (Km) values for Xyl6 and UDP-Xyl are 0.45 mM and 0.37 mM, respectively, as calculated from a Hanes-Woolf plot based on data presented in Fig 3E and 3F.
Fig 3. Biochemical properties of the xylan XylT activity in microsomes from garden asparagus spears.
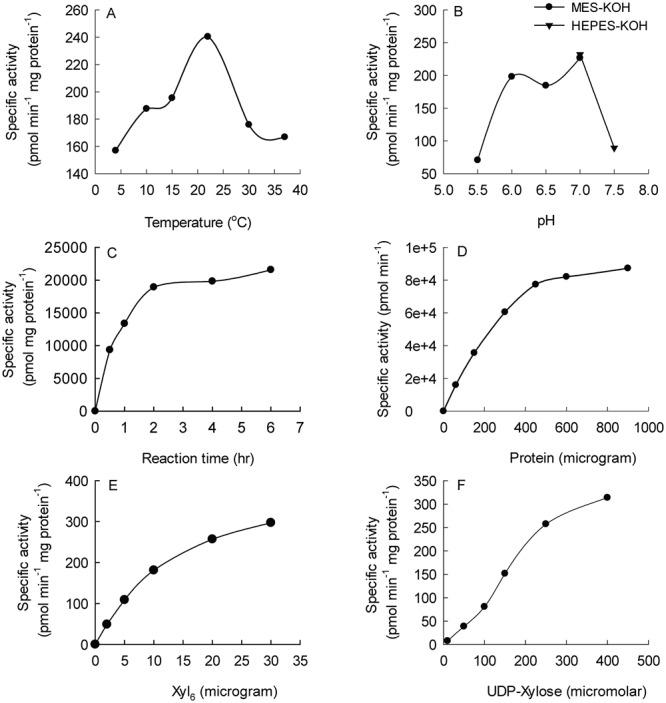
Microsomes extracted from the fresh basal Asparagus spears were incubated with UDP-[14C]-Xyl and Xyl6 for 1 hr unless otherwise indicated. The XylT activity was measured by counting the radioactivity at the origin of the paper chromatogram and expressed as specific activity. All assays were repeated twice and the values were averaged. Effect of temperature (A), pH (B), reaction time (C), microsomal protein amount (D), amount of Xyl6 (E) and UDP-Xyl concentration (F) on enzyme activity.
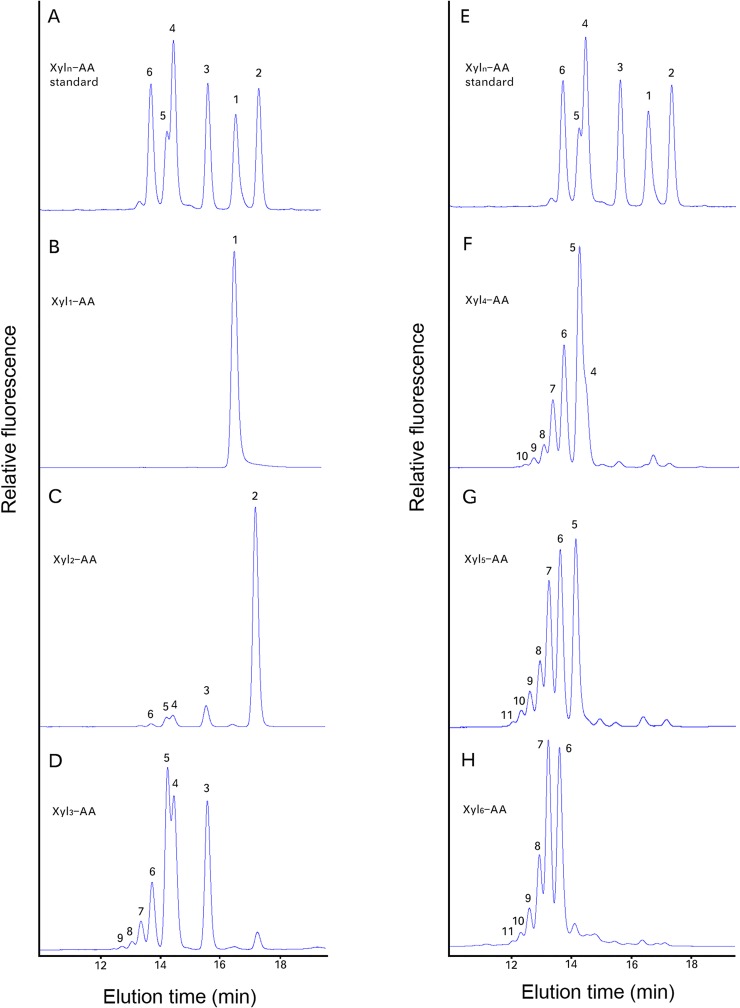
In order to more easily characterize the Asparagus products of the XylT activity assay, acceptor xylo-oligosaccharides were fluorescently tagged with anthranilic acid (AA) at their reducing end and used in a slightly modified XylT assay (see Materials and Methods). The assay products generated by Asparagus microsomes were then characterized by reversed-phase (RP)-HPLC (Figs 4 and 5), MALDI-TOF-MS (Fig 6) and enzyme digestion (Fig 7). Interestingly, the Xyl1-AA and Xyl2-AA were eluted in the “opposite” order to the rest of AA labeled xylo-oligosaccharide standards (DP3-6) (Fig 4A), which eluted faster with increasing DP. This order of the AA labeled xylo-oligosaccharide standard was confirmed by RP-HPLC-MS analysis (data not shown). No activity was detected with the monomer (Xyl1-AA) as the acceptor (Fig 4B). The Asparagus XylT activity increased with higher DP (2–6) of the acceptors (Fig 4C, 4D, 4F, 4G and 4H, respectively).
Fig 4. Xylan XylT activity measured using fluorescently tagged (AA) xylo-oligosaccharides of different lengths (Xyl1-Xyl6) as exogenous acceptors.
The reaction was conducted by mixing Asparagus microsomes with cold UDP-Xyl and the fluorescent acceptors Xyl1-AA (B), Xyl2-AA (C), Xyl3-AA (D), Xyl4-AA (F), Xyl5-AA (G) or Xyl6-AA (H) and incubated at RT for 1 hr. The reaction products were separated by RP-HPLC and detected by a fluorescence detector. A chromatogram showing a separation of a Xyl1-AA to Xyl6-AA mixture of standards is shown in (A) and (E) for reference. The numbers on the plots indicates the DP of the xylo-AA oligosaccharides.
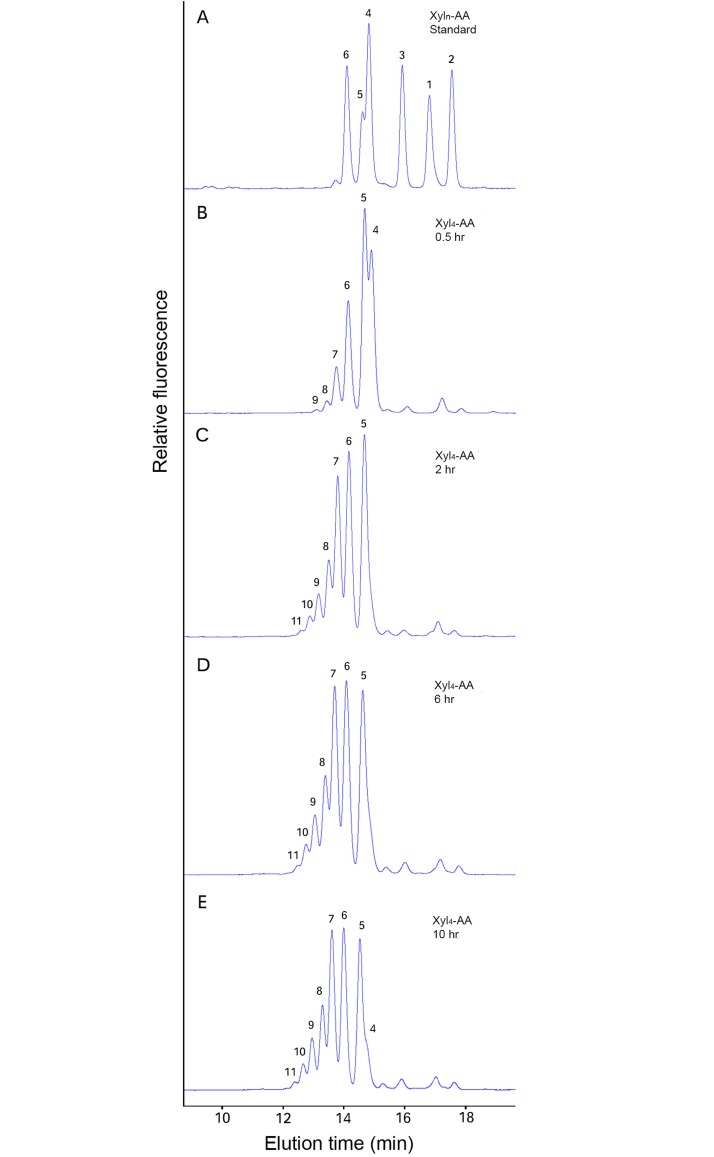
Fig 5. Time course of processive transfer of Xyl residues onto the Xyl4-AA acceptor.
Asparagus microsomes were incubated with UDP-Xyl and the Xyl4-AA acceptor for 0.5 hr (B), 2 hr (C), 6 hr (D) and 10 hr (E). The reaction products were analyzed by RP-HPLC. (A) Standard chromatogram of Xyl1-AA to Xyl6-AA.
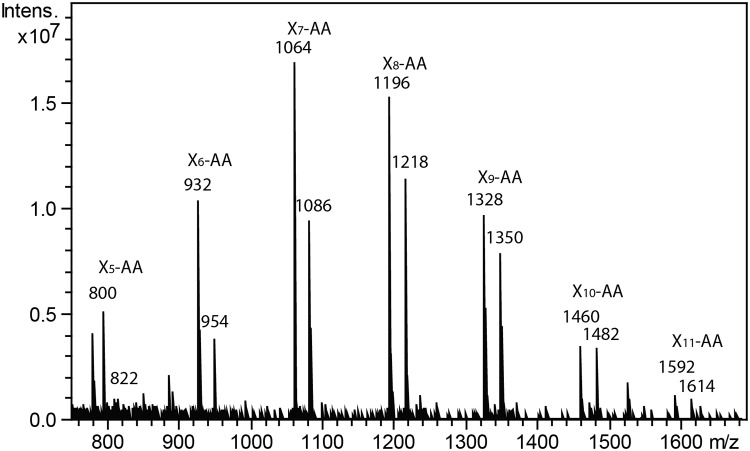
Fig 6. MALDI-TOF mass spectra of the xylan XylT reaction products catalyzed by Asparagus spear microsomes.
Microsomes were incubated with UDP-Xyl and the Xyl5-AA acceptor for 1 hr. Reaction products were purified by RP-HPLC and the fraction between 12 and 15 min collected and analyzed by MALDI-TOF MS. The ions corresponding to AA-labeled xylo-oligosaccharides (both in H+ and Na+ form) are labelled.
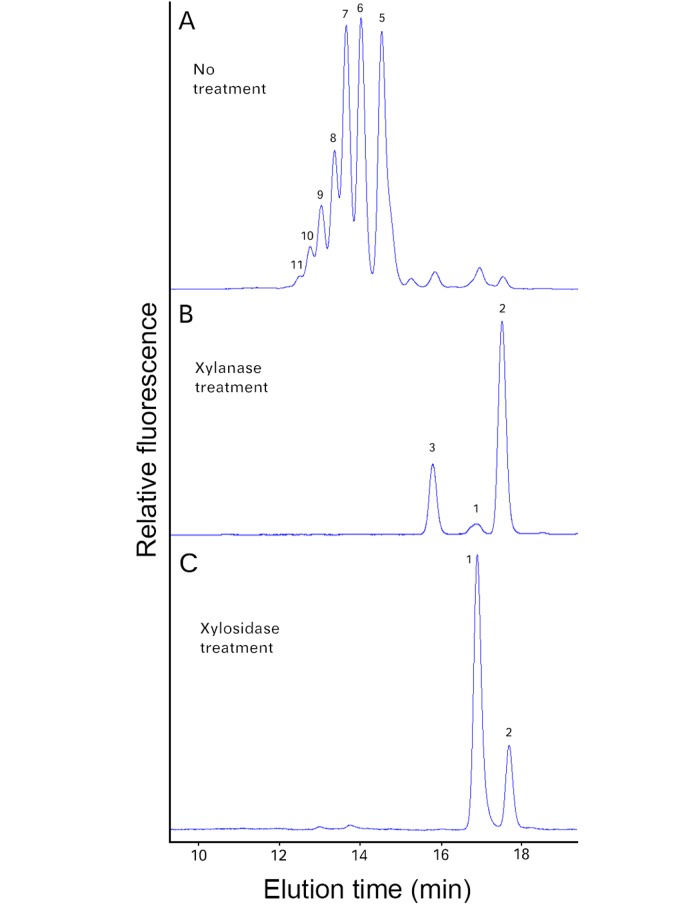
Fig 7. Enzymatic treatment of XylT reaction products generated using Xyl4-AA as acceptor.
Microsomes were incubated with UDP-Xyl and the Xyl4-AA acceptor, and the reaction products were then digested with endo-β-(1,4)-xylanase (B) and β-xylosidase (C). The reaction products with (B, C) and without (A) enzymatic digestion were analyzed by RP-HPLC.
Lee et al. [10]) demonstrated that in Arabidopsis a longer reaction time could lead to the production of higher DP xylo-oligosaccharides by the XylT. We also performed a time course study using Xyl4-AA as the acceptor (Fig 5) and found up to seven Xyl residues were added within a 2 hr incubation period, resulting in xylo-oligosaccharide products ranging from DP5-11 (Fig 5C). Longer incubation times, up to 10 hr, did not result in the formation of longer xylo-oligosaccharides (Fig 5).
The products of the XylT activity using Xyl5-AA as the acceptor were characterized by MALDI-TOF-MS. The observed molecular ion series (protonated and sodiated adducts) corresponded to the expected molecular masses for a pentitol series (+132 a.m.u) of oligosaccharides of DP 5–11 (m/z 800, 932, 1064, 1196, 1328, 1460, 1592 and 822, 954, 1086, 1218, 1350, 1482, 1614, respectively) series, corresponding to the H+ and Na+ forms, respectively (Fig 6).
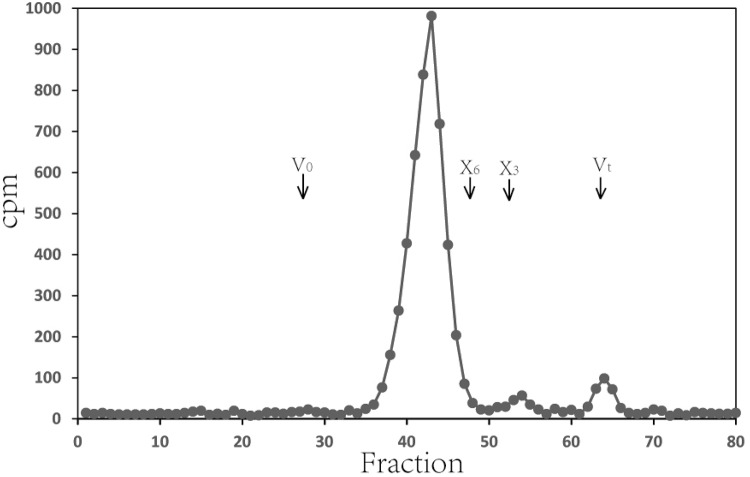
The products of the XylT reaction generated using Xyl6 as the acceptor and UDP-[14C]-Xyl as the donor were fractionated on a Bio-Gel P2 column (Fig 8). Around 5000 cpm of [14C]-Xyl product was loaded onto the column and the majority (96%) of the products were eluted between fraction 37 and 47, slightly earlier than the Xyl6 standard. There were two minor peaks at fractions 54 and 64 (Vt) corresponded to Xyl3 and Xyl1 respectively.
Fig 8. XylT reaction products generated using Xyl6 as the acceptor were fractionated on a Bio-Gel P2 column.
Asparagus microsomes were incubated with UDP-[14C]-Xyl and Xyl6 acceptor. The reaction products were treated with Dowex resin to remove unreacted UDP-[14C]-Xyl and fractionated on Bio-Gel P2 column. V0 and Vt, void and total volume, respectively. X6 and X3, xylo-hexaoligosaccharide and xylo-trioligosaccharide standards.
The linkage/anomeric configuration of the xylo-oligosaccharide products was then established by enzymatic hydrolysis using xylan-specific enzymes, namely an endo-β-(1,4)-xylanase and a β-xylosidase. The endo-xylanase degraded the xylo-oligosaccharides to DP2 and DP3 whereas the β-xylosidase degraded the xylo-oligosaccharides to DP1 and some DP2 (Fig 7), confirming that the Xyl is linked β–(1,4).
A comparison of the xylan XylT activity in garden asparagus spears during post-harvest cold storage
To investigate changes in xylan XylT activity during the storage process, Asparagus spear tissues from six stages (0, 2, 4, 8, 12, 14 days post-harvest) were collected and the XylT activity of the apical, middle and basal sections was analyzed. The basal tissue was found to have four times higher XylT activity than the apical section in freshly harvested tissues, correlating with the increased heteroxylan content of this section compared to the apex (Fig 9). Notably, the XylT activity slightly increased during the initial stage of storage (0–2 day after harvest) and then declined in all three sections with additional storage time (Fig 9).
Fig 9. Xylan XylT activities of different Asparagus sections over the 4°C storage period.
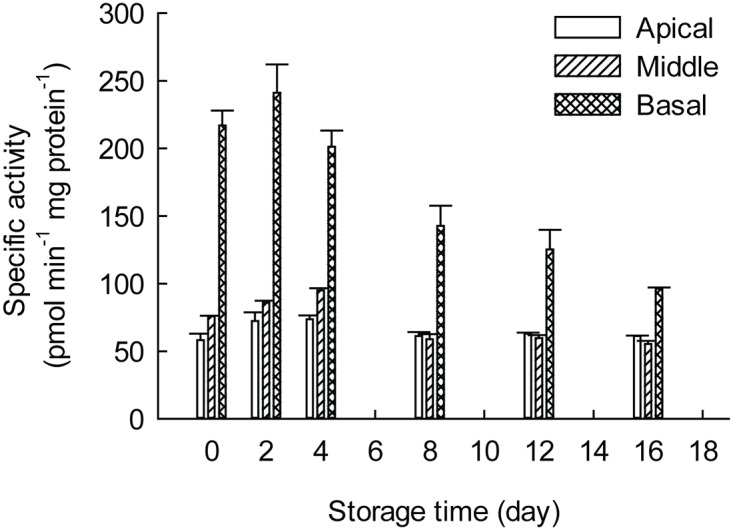
Microsomes from apical, middle and basal sections of Asparagus spears stored at 4°C for 0–16 days were isolated and the XylT activities were measured as described in the Materials and Methods. Data were average values±SE (n = 3).
The Asparagus spear offers distinct advantages to study the biochemical/molecular mechanism(s) of xylan biosynthesis with its vastly superior xylan XylT activity compared to other plant models, yet Asparagus officinalis lacks a sequenced genome. We therefore decided to use an RNA-Seq approach to identify the genes involved in xylan biosynthesis in this system. In an initial experiment, a combined sample of total RNA extracted from fresh and 4 day-old cold-stored Asparagus spears was sequenced. The raw RNA-Seq dataset comprised 191,179,245 paired-end reads. After quality trimming and adapter removal, a total of 174,487,864 read pairs remained and were assembled into 403,090 contigs (N50 47503, L50 1824bp, 293.4Mb). The assembly contigs were then scaffolded [44] which resulted in a reference transcriptome consisting of 351,350 transcript isoforms.
BLASTX searches were used to identify the putative Asparagus orthologs of known Arabidopsis xylan synthesis genes including GTs (IRX9, IRX10, IRX14, FRA8, F8H, IRX8, PARVUS, GUX1) and non-GTs (UXS, GXM, IRX15, TBL29). Other secondary wall biosynthesis genes including phenylalanine ammonia-lyase (PAL) [49] and the NAC secondary wall thickening promoting factor-1 (NST1) [50] representing proteins involved in lignin biosynthesis and a secondary wall transcription factor, respectively, were also searched within the reference transcriptome. As the focus of this study was on xylan XylT activity, IRX9, IRX10 and IRX14 and their related homologs were chosen for further analysis. The full-length open reading frames of five XylT genes were identified and designated AoIRX9, AoIRX9-L, AoIRX10, AoIRX14_A, AoIRX14_B on the basis of their similarity to the Arabidopsis sequences. The corresponding contigs are listed in Table 2. IRX9 and IRX14 belong to CAZy family GT43 (http://www.cazy.org/) and a phylogenetic tree was created using Arabidopsis and Asparagus GT43 protein sequences (S1 Fig). Both Arabidopsis IRX9 and IRX-9L have apparent orthologs in Asparagus, although it is uncertain which Asparagus sequence is the true ortholog in the case of IRX-14_A/14_B, hence their generic naming. Interestingly, only one IRX10 gene was found in the Asparagus transcriptome which shares 86% and 89% identity with AtIRX10 and AtIRX10-L, respectively (S2 Fig). The full-length sequences of these genes have been deposited in the Genbank database (http://www.ncbi.nlm.nih.gov/Genbank) with the following accession numbers: AoIRX9 (KJ556998); AoIRX9-L (KJ556999); AoIRX10 (KJ557000); AoIRX14_A (KJ557001); AoIRX14_B (KJ557002).
Table 2. Asparagus orthologs of Arabidopsis xylan biosynthetic genes as well as GT2 (cellulose synthase and cellulose synthase like genes) and representatives of known secondary wall associated genes.
| Asparagus contig no. | AGI | Gene Name | Proposed Function | E-Value | Stem section | ||
|---|---|---|---|---|---|---|---|
| Apical | Middle | Basal | |||||
| comp82040_c0 | AT2G37090 | IRX9 | backbone synthesis | e-103 | 0.17±0.02 | 9.93±0.32 | 13.73±0.18 |
| comp85515_c0 | AT1G27600 | IRX9-L | backbone synthesis | e-105 | 55.13±1.48 | 48.58±2.48 | 38.76±1.52 |
| comp90084_c0 | AT1G27440 | IRX10 | backbone synthesis | 0 | 40.05±2.15 | 44.87±1.67 | 43.43±0.77 |
| comp84164_c0 | AT5G67230 | IRX14-L | backbone synthesis | e-131 | 19.26±1.23 | 21.90±0.59 | 22.1±1.11 |
| comp86147_c0 | AT5G67230 | IRX14-L | backbone synthesis | e-102 | 14.47±0.98 | 23.61±0.86 | 19.82±0.34 |
| comp89578_c0 | AT2G28110 | IRX7 | reducing end synthesis | e-145 | 28.67±1.02 | 35.87±0.89 | 52.76±2.76 |
| comp84659_c0 | AT5G22940 | IRX7-L | reducing end synthesis | 0 | 1.84±0.10 | 3.75±0.26 | 3.89±0.09 |
| comp29843_c0 | AT5G54690 | IRX8 | reducing end synthesis | 0 | 5.64±0.40 | 6.67±0.34 | 7.86±0.54 |
| comp74252_c0 | AT5G54690 | IRX8 | reducing end synthesis | 0 | 7.23±0.35 | 7.35±0.62 | 8.87±0.66 |
| comp88939_c1 | AT1G19300 | PARVUS | reducing end synthesis | e-155 | 40.79±2.25 | 45.9±4.44 | 57.07±4.28 |
| comp84705_c0 | AT3G18660 | GUX1 | GlcA incorporation | 0 | 18.46±0.70 | 14.39±0.82 | 15.87±0.36 |
| comp109454_c0 | AT3G18660 | GUX1 | GlcA incorporation | e-122 | 2.25±0.76 | 4.36±1.09 | 5.06±0.71 |
| comp79489_c0 | AT3G62830 | UXS2 | UDP-Xyl synthase | 0 | 75.6±3.86 | 51.41±3.32 | 45.58±2.40 |
| comp61026_c0 | AT3G53520 | UXS1 | UDP-Xyl synthase | e-171 | 53.58±1.50 | 53.11±1.86 | 59.72±0.83 |
| comp37556_c0 | AT1G09610 | GXM1 | GlcA methyltransferase | 3e-91 | 2.69±0.30 | 11.6±0.58 | 10.82±0.72 |
| comp60971_c0 | AT5G67210 | IRX15-L | unknown | 4e-67 | 10.15±1.77 | 4.1±0.41 | 4.26±1.08 |
| comp80607_c1 | AT3G55990 | TBL29 | acetyltransferase | 0 | 0.18±0.05 | 1.5±0.18 | 1.07±0.23 |
| comp67577_c0 | AT3G55990 | TBL29 | acetyltransferase | e-158 | 0.52±0.12 | 2.13±0.46 | 1.66±0.21 |
| comp91651_c0 | AT4G32410 | CESA1 | cellulose synthase | 0 | 13.31±0.76 | 15.80±1.42 | 13.35±1.26 |
| comp82725_c0 | AT5G05170 | CESA3 | cellulose synthase | 0 | 4.16±0.41 | 4.31±0.29 | 4.24±0.33 |
| comp88863_c0 | AT5G44030 | CESA4 | cellulose synthase | 0 | 27.06±1.32 | 43.67±0.96 | 39.05±1.97 |
| comp86858_c0 | AT5G09870 | CESA5 | cellulose synthase | 0 | 0.82±0.18 | 1.70±0.22 | 1.84±0.25 |
| comp90588_c1 | AT5G64740 | CESA6 | cellulose synthase | 0 | 230.2±6.44 | 291.1±35.4 | 247.9±36.6 |
| comp43060_c0 | AT5G17420 | CESA7 | cellulose synthase | 0 | 0.39±0.11 | 11.76±0.51 | 13.91±0.69 |
| comp76272_c0 | AT5G03760 | CSLA9 | mannan synthase | 0 | 11.77±0.26 | 13.84±1.25 | 16.24±0.90 |
| comp78814_c0 | AT5G03760 | CSLA9 | mannan synthase | 0 | 10.03±0.54 | 10.33±0.47 | 14.93±0.81 |
| comp78728_c1 | AT5G03760 | CSLA9 | mannan synthase | e-175 | 13.59±0.15 | 8.01±0.38 | 14.39±0.81 |
| comp187879_c0 | AT5G03760 | CSLA9 | mannan synthase | 0 | 0.28±0.10 | 1.14±0.36 | 0.95±0.21 |
| comp80676_c0 | AT4G07960 | CSLC12 | xyloglucan synthase | 0 | 30.20±1.48 | 18.71±1.78 | 17.57±0.52 |
| comp83916_c0 | AT3G03050 | CSLD3 | cellulose synthase | 0 | 17.64±1.00 | 16.55±2.02 | 18.69±1.66 |
| comp87435_c0 | AT1G55850 | CSLE1 | unknown | 0 | 17.78±1.30 | 16.73±1.35 | 24.04±1.52 |
| comp89194_c0 | AT1G55850 | CSLE1 | unknown | e-179 | 34.80±0.84 | 32.19±1.61 | 29.54±2.44 |
| comp81970_c0 | AT4G23990 | CSLG3 | unknown | e-179 | 13.95±0.39 | 18.46±0.86 | 26.05±1.82 |
| comp86723_c0 | AT2G37040 | PAL | lignin synthesis | 0 | 28.09±1.01 | 41.67±2.48 | 127.73±2.78 |
| comp74161_c0 | AT2G46770 | NST1 | secondary wall TF | 2e-84 | 0.33±0.12 | 2.53±0.38 | 3.54±0.43 |
| comp59563_c0 | AT5G09810 | ACTIN | house keeping | 0 | 936.3±15.3 | 943.1±25.7 | 844.1±31.6 |
Gene expression in the basal, middle and apical sections is shown as transcripts per million (TPM) ± SD (n = 4). The house keeping gene ACTIN is used a control.
A separate quantitative RNA-Seq experiment was also conducted using four biological replicates of apical, middle and basal section from fresh Asparagus spears. The relative expression values (transcripts per million reads, TPM) of the five Asparagus putative xylan backbone XylT genes in the basal, middle and apical stem sections are listed in Table 2. AoIRX9-L, AoIRX10, AoIRX14_A and AoIRX14_B do not display differential expression across the spear sections, in contrast to AoIRX9 which shows substantially higher expression in the middle and basal stem sections compared to the apex. This expression pattern is similar to AoPAL and AoNST1 whose expression is higher in the base of the stem where more secondary wall synthesis is occurring.
The expression of the five xylan backbone XylT genes was also analyzed by semi-quantitative RT-PCR to confirm the pattern and relative transcript abundance observed in RNA-Seq analysis. The expression of AoIRX9 in the basal and middle sections was again observed to be higher than that of the apical section while other xylan backbone genes show no differential expression throughout all three sections of the spear (S3 Fig). As expected, PAL and NST1 were more highly expressed in the basal compared to the apical section. In all cases, the pattern and intensity of the amplified PCR product correlated well with the RNA-Seq quantitation results, providing robust validation of these data (Table 2).
Discussion
Asparagus spears undergo significant physiological and biochemical changes after harvest, including increased lignification
As an immature and fast growing stem tissue, Asparagus spears retain high physiological activities including high respiration rates, the depletion of soluble sugars, proteins and ascorbic acid, and pronounced water losses after harvest and cold storage in the dark [36, 51]. In the present study, the spears rapidly lost water content and their visual appearance was reduced to the point of being unattractive to consumers after 16 days of storage at 4°C (Fig 1A and 1B). The more mature basal part of the fresh spear had a harder texture and higher content of lignin than the immature apical part (Fig 1C and 1D). The continuation of shoot differentiation after harvest results in secondary wall thickening and increased lignification. However, the process of hardening does not take place uniformly along the length of the spear due to differing respiration rates [36]. Significant increases in lignin levels were found in all three sections upon cold storage, with the greatest increase observed in the basal section (Fig 1D), indicating cold treatment-induced lignification of garden asparagus spears. However, there were no significant changes in texture in either the middle or apical sections during cold storage, whereas the texture of the basal section was the greatest in the sections at day 0, and increased until day 8 of storage and then decreased to the day 0 level by day 16 (Fig 1C). These results are consistent with those described by Villanueva-Suarez et al. [52] who suggested the hardening process was related to wall polysaccharide changes and water loss as well as the amounts of lignin. Further, our data (see Discussion below) would suggest that the process likely also involves the relative lowly substituted heteroxylans that increase significantly during this period of storage (Table 1 and S1 Table) and which would be expected to self-associate to form “hydrophobic bundles” of insoluble polysaccharides that exclude water, much like cellulose microfibrils adding further rigidity to the wall[53,54].
An in-vitro xylan XylT assay indicates Asparagus spears actively synthesize heteroxylan during storage at 4°C
Previous reports of xylan XylT activity have mostly focused on tissues of dicot and commelinoid monocot species undergoing normal growth and development but there is scant information about changes in XylT activity for non-commelinoid monocot species such as Asparagus. We detected a xylan XylT activity in the apical, middle and basal sections of Asparagus spears that increases during the early phase of cold storage after harvest and then decreases (Fig 9). The increased XylT activity was coincident with the lignification of Asparagus spears, suggesting a coordinated deposition of secondary wall polymers resulting in a rigidification of the spears. Using a biochemical assay based upon the incorporation of Xyl from UDP-Xyl into a xylo-oligosaccharide acceptor, we showed that Asparagus spear xylan XylT activity is significantly higher (thirteen fold) than in Arabidopsis stems (Fig 3). As a non-commelinoid monocot and different from grasses, Asparagus has a type-I cell wall composition similar to dicots [55,56].
We have demonstrated that the Asparagus XylT has processive activity capable of adding up to seven Xyl residues to a xylo-oligosaccharide acceptor of DP4 (see Fig 5) with kinetic characteristics (optimal pH, temperature, etc.) similar to those previous published in Arabidopsis [10] and wheat [27]. The Km values of UDP-Xyl and pyridylaminated (PA) Xyl5 (Xyl6-PA was not analyzed) of etiolated wheat seedlings were 7.9mM and 2.9mM [27], respectively, higher than what was observed for the Asparagus microsomes. This correlates with the higher XylT activity of Asparagus. The Km value of Xyl6 (0.45 mM) is lower than previous published for the purified AtIRX10-L protein (1.17 mM) [21] but this could be due to a different combination of proteins in the xylan XylS in the biosynthetic protein complex of Asparagus compared to Arabidopsis. However, the Asparagus XylS is more amenable for purification due to its considerably higher activity thus assisting in studying its molecular mechanism of action.
The degree of polymerization (DP) of xylo-oligosaccharides synthesized by Asparagus microsomes in vitro (DP 7–11) is still far smaller than the native heteroxylan, which is between 150–200 residues [36]. It is generally assumed that the “xylan synthase (XylS)” in vivo is a protein complex comprising multiple GTs [5], although the mechanism that controls the length of the xylan chain and the termination of xylan biosynthesis is not well understood [5]. Based upon assays of heterologously expressed IRX proteins and in vitro biochemical assays, Urbanowicz et al [21] have proposed that the Arabidopsis IRX10-L is the catalytic xylan synthase (XYLAN SYNTHASE-1, XYL1) and whilst not ruling out a catalytic function for IRX 9/14 they propose that although these two proteins are a part of the xylan synthase complex their role is more likely in either chain initiation and/or a structural role in stabilizing the synthase complex.
Next-generation sequencing provides a powerful tool to use Asparagus as a model to study heteroxylan biosynthesis
The recently developed RNA-seq technology is a powerful tool to study gene expression, especially for non-model species [57]. The Asparagus RNA-seq dataset makes a genome wide comparison between Asparagus and the model dicot Arabidopsis possible. It is not surprising that all the Arabidopsis xylan biosynthesis related genes have orthologs in Asparagus (Table 2). Arabidopsis IRX9(-L) and IRX14(-L) belong to the CAZy GT43 family. Arabidopsis mutant studies show that they are separate classes of two non-redundant genes both involved in xylan biosynthesis and that IRX9 and IRX14 play the major role while the redundant IRX9-L and IRX14-L function in a minor role [11]. Tobacco suspension cells co-expressing the Arabidopsis IRX9 and IRX14 have been shown to possess xylan backbone elongation activity but the activity is absent when either gene is expressed alone [28]. This suggests that IRX9 and IRX14 operate in a xylan biosynthesis protein complex. In grasses, rice IRX9 and IRX14 have been demonstrated to be involved in xylan biosynthesis via Arabidopsis mutant complementation analyses [58]. In this study, AoIRX9 shows a distinct expression pattern compared with the other IRX genes. Its expression is significantly higher in the basal section than in the apical section in fresh Asparagus, and this correlates with the higher xylan XylT catalytic activity in the basal tissue. The semi-quantitative RT-PCR data is consistent with RNA-seq analysis of the gene expression in the basal, middle and apical sections of fresh Asparagus spears (Table 2 and S3 Fig). A site-directed mutagenesis study on AtIRX9 has shown that IRX9 appears not to be directly involved in catalytic activity [59]. Combined with the quantitative RNA-seq analysis, this indicates that AoIRX9 is playing a key role in regulating xylan biosynthesis during secondary wall biosynthesis in Asparagus. For example, AoIRX9 may function by recruiting the other XylTs to form an active xylan synthase complex in the Golgi apparatus, consistent with the conclusions drawn by Urbanowicz et al [21].
Mutant analysis has revealed Arabidopsis IRX10 and IRX10-L play an essential role in xylan biosynthesis. The two homologs are functionally redundant and double mutants show a severe growth phenotype and are also infertile [8,9]. Down-regulation of rice OsIRX10 leads to decreased xylan and a moss IRX10 could only partially complement the Arabidopsis irx10 irx10-L double mutant [60,61]. Urbanowicz et al [21] have provided biochemical evidence that AtIRX10-L (XYL1) is a xylan synthase while no XylT was demonstrated for AtIRX9 and AtIRX14. Although IRX10 protein sequences are conserved from mosses to vascular plants (S2 Fig) and it remains unknown what role IRX10 plays in the non-vascular plant. Interestingly, only one IRX10 gene was identified in the Asparagus spear transcriptome. A blast analysis of the Asparagus densiflorus transcriptome (1000 plants initiative, https://sites.google.com/a/ualberta.ca/onekp/) with Arabidopsis IRX10 also identified only one IRX10 (data not shown), providing further evidence that there may only be one IRX10 gene in Asparagus species although sequencing of the Asparagus genome will be required to provide a definitive answer. It still remains a mystery as to how IRX9, IRX10 and IRX14 interact with each other (and other GTs and the O-acetyl transferase (XOAT1)) to coordinate heteroxylan backbone synthesis.
Genomic analysis suggests that the Asparagales are more similar to eudicots than to Poales [62]. Given most species in this order have large genomes, the genetic information currently available for these non-commelinoid monocots is limited. The family Asparagaceae has the smallest genome among the Asparagales and it has been proposed that Asparagus be used as a genomic model [63]. In this study, the transcriptome database provides an essential tool to study the gene expression and characterization of candidate genes/proteins associated with xylan synthesis in Asparagus.
Conclusion
We have identified and characterized a highly active xylan XylT in Asparagus officinalis, a non-commelinoid monocot. This XylT activity correlates with increased xylan content and is the most promising plant model in which it is sufficiently active in in vitro microsomal preparations to enable purification of the native xylan synthase protein complex (XylS). Transcriptome analysis revealed five candidate XylT synthase genes which together with a functional XylT assay will allow us to establish their precise roles in xylan biosynthesis and shed new light on the mechanism of assembly.
Supporting Information
Four predicted Asparagus sequences (AoIRX9, AoIRX9-L, AoIRX14_A and AoIRX14_B) were aligned with Arabidopsis GT43 proteins using Clustal X 2.1 and a neighbor-joining tree was produced. The bar represents a percent accepted mutations value of 10%. The numbers shown at branching points are bootstrap values derived from 1000 randomized samples.
(TIF)
IRX10 sequences were aligned using Clustal X 2.1. Alignment shown in Jalview 2.8 where blue shading indicates sequence identity across the five sequences. Greater than 80% identity is dark blue, greater than 60% is medium blue, greater than 40% is light blue and 40% or less has no shading.
(TIF)
Total RNA was extracted from apical, middle and basal sections of fresh Asparagus spears and the expression patterns of AoIRX9, AoIRX9-L, AoIRX10, AoIRX14_A, AoIRX14_B, PAL and NST1, were analyzed by semi-quantitative RT-PCR using actin as a control.
(TIF)
(DOC)
(DOC)
Acknowledgments
We thank Mr. Sybrand Visagie from Vizzarri farm, Melbourne, Victoria, for providing the Asparagus samples and Dr Berin Boughton for MALDI-TOF analysis.
Data Availability
The full-length sequences of these genes are available from the Genbank database (http://www.ncbi.nlm.nih.gov/Genbank) with the following accession numbers: AoIRX9 (KJ556998); AoIRX9-L (KJ556999); AoIRX10 (KJ557000); AoIRX14_A(KJ557001); AoIRX14_B (KJ557002).
Funding Statement
This work was funded by a grant from the Australia Research Council to the ARC Centre of Excellence in Plant Cell Walls (WZ, KP, EL, CB, AC, AL, MSD and AB) (CE110001007); ARC DP110100410 (EL and AB), the Victorian Life Sciences Computation Initiative (VLSCI) grant number VR0191 on its Peak Computing Facility at the University of Melbourne, an initiative of the Victorian Government, Australia and the National Natural Science Foundation of China (LS) (grant no. 31170165). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Doblin MS, Pettolino F, Bacic A. Plant cell walls: the skeleton of the plant world. Funct Plant Biol. 2010; 37: 357–381. [Google Scholar]
- 2. Faik A. Xylan biosynthesis: news from the grass. Plant Physiol. 2010; 153: 396–402. 10.1104/pp.110.154237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ebringerova A, Heinze T. Xylan and xylan derivatives—biopolymers with valuable properties, 1—Naturally occurring xylans structures, procedures and properties. Macromol Rapid Comm. 2000; 21: 542–556. [Google Scholar]
- 4. Bacic A, Stone B. A (1–3)-linked and (1–4)-linked beta-D-glucan in the endosperm cell-walls of wheat. Carbohyd Res. 1980; 82: 372–377. [Google Scholar]
- 5. Rennie EA, Scheller HV. Xylan biosynthesis. Curr Opin Biotech. 2014; 26: 100–107. 10.1016/j.copbio.2013.11.013 [DOI] [PubMed] [Google Scholar]
- 6. Hao Z, Mohnen D. A review of xylan and lignin biosynthesis: Foundation for studying Arabidopsis irregular xylem mutants with pleiotropic phenotypes. Criti Rev Biochemi Mol Biol. 2014; 49: 212–241. 10.3109/10409238.2014.889651 [DOI] [PubMed] [Google Scholar]
- 7. Zhong RQ, Pena MJ, Zhou GK, Nairn CJ, Wood-Jones A, et al. Arabidopsis fragile fiber8, which encodes a putative glucuronyltransferase, is essential for normal secondary wall synthesis. Plant Cell. 2005; 17: 3390–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu AM, Rihouey C, Seveno M, Hornblad E, Singh SK, Matsunaga T, et al. The Arabidopsis IRX10 and IRX10-LIKE glycosyltransferases are critical for glucuronoxylan biosynthesis during secondary cell wall formation. Plant J. 2009; 57: 718–731. 10.1111/j.1365-313X.2008.03724.x [DOI] [PubMed] [Google Scholar]
- 9. Brown DM, Zhang ZN, Stephens E, Dupree P, Turner SR. Characterization of IRX10 and IRX10-like reveals an essential role in glucuronoxylan biosynthesis in Arabidopsis. Plant J. 2009; 57: 732–746. 10.1111/j.1365-313X.2008.03729.x [DOI] [PubMed] [Google Scholar]
- 10. Lee CH, O'Neill MA, Tsumuraya Y, Darvill AG, Ye ZH. The irregular xylem9 mutant is deficient in xylan xylosyltransferase activity. Plant Cell Physiol. 2007; 48: 1624–1634. [DOI] [PubMed] [Google Scholar]
- 11. Wu AM, Hornblad E, Voxeur A, Gerber L, Rihouey C, Lerouge P, et al. Analysis of the Arabidopsis IRX9/IRX9-L and IRX14/IRX14-L pairs of glycosyltransferase genes reveals critical contributions to biosynthesis of the hemicellulose glucuronoxylan. Plant Physiol. 2010; 153: 542–554. 10.1104/pp.110.154971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pena MJ, Zhong RQ, Zhou GK, Richardson EA, O'Neill MA, Darvill AG, et al. Arabidopsis irregular xylem8 and irregular xylem9: Implications for the complexity of glucuronoxylan biosynthesis. Plant Cell. 2007; 19: 549–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee CH, Zhong RQ, Richardson EA, Himmelsbach DS, McPhail BT, Ye ZH. The PARVUS gene is expressed in cells undergoing secondary wall thickening and is essential for glucuronoxylan biosynthesis. Plant Cell Physiol. 2007; 48: 1659–1672. [DOI] [PubMed] [Google Scholar]
- 14. Mortimer JC, Miles GP, Brown DM, Zhang ZN, Segura MP, Weimar T, et al. Absence of branches from xylan in Arabidopsis gux mutants reveals potential for simplification of lignocellulosic biomass. P Natl Acad Sci USA. 2010; 107: 17409–17414. 10.1073/pnas.1005456107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brown DM, Goubet F, Vicky WWA, Goodacre R, Stephens E, Dupree P, et al. Comparison of five xylan synthesis mutants reveals new insight into the mechanisms of xylan synthesis. Plant J. 2007; 52: 1154–1168. [DOI] [PubMed] [Google Scholar]
- 16. Oikawa A, Joshi HJ, Rennie EA, Ebert B, Manisseri C, Heazlewood JL, et al. An integrative approach to the identification of Arabidopsis and rice genes involved in xylan and secondary wall development. Plos One. 2010; 5: e15481 10.1371/journal.pone.0015481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee C, Teng Q, Huang WL, Zhong RQ, Ye ZH. The Arabidopsis family GT43 glycosyltransferases form two functionally nonredundant groups essential for the elongation of glucuronoxylan backbone. Plant Physiol. 2010; 153: 526–541. 10.1104/pp.110.155309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee C, Teng Q, Zhong RQ, Yuan YX, Haghighat M, Ye ZH. Three Arabidopsis DUF579 domain-containing GXM proteins are methyltransferases catalyzing 4-O-methylation of glucuronic acid on xylan. Plant Cell Physiol. 2012; 53: 1934–1949. 10.1093/pcp/pcs138 [DOI] [PubMed] [Google Scholar]
- 19. Piston F, Uauy C, Fu LH, Langston J, Labavitch J, Dubcovsky J. Down-regulation of four putative arabinoxylan feruloyl transferase genes from family PF02458 reduces ester-linked ferulate content in rice cell walls. Planta. 2010; 231: 677–691. 10.1007/s00425-009-1077-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xiong GY, Cheng K, Pauly M. Xylan O-acetylation impacts xylem development and enzymatic recalcitrance as indicated by the arabidopsis mutant tbl29. Mol Plant. 2013; 6: 1373–1375. 10.1093/mp/sst014 [DOI] [PubMed] [Google Scholar]
- 21. Urbanowicz BR, Pena MJ, Moniz HA, Moremen KW, York WS. Two Arabidopsis proteins synthesize acetylated xylan in vitro . Plant J. 2014; 80: 197–206. 10.1111/tpj.12643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Feingold DS, Neufeld EF, Hassid WZ. Xylosyl transfer catalyzed by an asparagus extract. J Biol Chem. 1959; 234: 488–489. [PubMed] [Google Scholar]
- 23. Bailey RW, Hassid WZ. Xylan synthesis from uridine-diphosphate-D-xylose by particulate preparations from immature corncobs. P Natl Acad Sci USA. 1966; 56: 1586–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dalessandro G, Northcote DH. Increase of xylan synthetase-activity during xylem differentiation of the vascular cambium of sycamore and poplar trees. Planta. 1981; 151: 61–67. 10.1007/BF00384238 [DOI] [PubMed] [Google Scholar]
- 25. Waldron KW, Brett CT. A glucuronyltransferase involved in glucuronoxylan synthesis in pea (Pisum sativum) epicotyls. Biochem J. 1983; 213: 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Suzuki K, Ingold E, Sugiyama M, Komamine A. Xylan synthase activity in isolated mesophyll-cells of Zinnia elegans during differentiation to tracheary elements. Plant Cell Physiol. 1991; 32: 303–306. [Google Scholar]
- 27. Kuroyama H, Tsumuraya Y. A xylosyltransferase that synthesizes beta-(1 -> 4)-xylans in wheat (Triticum aestivum L.) seedlings. Planta. 2001; 213: 231–240. [DOI] [PubMed] [Google Scholar]
- 28. Lee C, Zhong RQ, Ye ZH. Arabidopsis family GT43 members are xylan xylosyltransferases required for the elongation of the xylan backbone. Plant Cell Physiol. 2012; 53: 135–143. 10.1093/pcp/pcr158 [DOI] [PubMed] [Google Scholar]
- 29. Zeng W, Jiang N, Nadella R, Killen TL, Nadella V, Faik A. A glucurono(arabino)xylan synthase complex from wheat contains members of the GT43, GT47, and GT75 families and functions cooperatively. Plant Physiol. 2010; 154: 78–97. 10.1104/pp.110.159749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jensen JK, Johnson NR, Wilkerson CG. Arabidopsis thaliana IRX10 and two related proteins from psyllium and Physcomitrella patens are xylan xylosyltransferases. Plant J. 2014; 80: 207–215. 10.1111/tpj.12641 [DOI] [PubMed] [Google Scholar]
- 31. Johansson MH, Samuelson O. Reducing end groups in birch xylan and their alkaline-degradation. Wood Sci Technol. 1977; 11: 251–263. [Google Scholar]
- 32. Ratnayake S, Beahan CT, Callahan DL, Bacic A. The reducing end sequence of wheat endosperm cell wall arabinoxylans. Carbohydr Res. 2013; 386C: 23–32. [DOI] [PubMed] [Google Scholar]
- 33. Mazumder K, York WS. Structural analysis of arabinoxylans isolated from ball-milled switchgrass biomass. Carbohyd Res. 2010; 345: 2183–2193. 10.1016/j.carres.2010.07.034 [DOI] [PubMed] [Google Scholar]
- 34. Haard NF, Sharma SC, Wolfe R, Frenkel. Ethylene induced isoperoxidase changes during fiber formation in postharvest asparagus. J Food Sci, 1974; 39: 452–456. [Google Scholar]
- 35. Waldron KW, Selvendran RR. Effect of maturation and storage on asparagus (Asparagus officinalis) cell-wall composition. Physiol Plantarum. 1990; 80: 576–583. [Google Scholar]
- 36. Waldron KW, Selvendran RR. Cell-wall changes in immature asparagus stem tissue after excision. Phytochemistry. 1992; 31: 1931–1940. [DOI] [PubMed] [Google Scholar]
- 37. Tzoumaki MV, Biliaderis CG, Vasilakakis M. Impact of edible coatings and packaging on quality of white asparagus (Asparagus officinalis, L.) during cold storage. Food Chem. 2009; 117: 55–63. [Google Scholar]
- 38. Pettolino FA, Walsh C, Fincher GB, Bacic A. Determining the polysaccharide composition of plant cell walls. Nat Protoc. 2012; 7: 1590–1607. 10.1038/nprot.2012.081 [DOI] [PubMed] [Google Scholar]
- 39. Luo ZS, Xu XL, Yan BF. Accumulation of lignin and involvement of enzymes in bamboo shoot during storage. Eur Food Res Technol. 2008; 226: 635–640. [Google Scholar]
- 40. Zeng W, Chatterjee M, Faik A. UDP-xylose-stimulated glucuronyltransferase activity in wheat microsomal membranes: Characterization and role in glucurono(arabino) xylan biosynthesis. Plant Physiol. 2008; 147: 78–91. 10.1104/pp.107.115576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ishikawa M, Kuroyama H, Takeuchi Y, Tsumuraya Y. Characterization of pectin methyltransferase from soybean hypocotyls. Planta. 2000; 210: 782–791. [DOI] [PubMed] [Google Scholar]
- 42. Alwael H, Connolly D, Paull B. Liquid chromatographic profiling of monosaccharide concentrations in complex cell-culture media and fermentation broths. Anal Methods-Uk. 2011; 3: 62–69. [DOI] [PubMed] [Google Scholar]
- 43. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011; 29: 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huang XQ, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999; 9: 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011; 12: 323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BM, et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013; 29: 1035–1043. 10.1093/bioinformatics/btt087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007; 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- 48. Rodriguez R, Jimenez A, Guillen R, Heredia A, Fernandez-Bolanos J. Postharvest changes in white asparagus cell wall during refrigerated storage. J Agr Food Chem. 1999; 47: 3551–3557. [DOI] [PubMed] [Google Scholar]
- 49. Rasmussen S, Dixon RA. Transgene-mediated and elicitor-induced perturbation of metabolic channeling at the entry point into the phenylpropanoid pathway. Plant Cell. 1999; 11: 1537–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhong RQ, Demura T, Ye ZH. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell. 2006; 18: 3158–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Papadopoulou PP, Siomos AS, Dogras CC. Textural and compositional changes of green and white asparagus spears during storage. Acta Hortic. 2002; 579: 647–651. [Google Scholar]
- 52. Villanueva-Suarez MJ, Redondo-Cuenca A, Rodriguez-Sevilla MD, Heredia-Moreno A. Postharvest storage of white asparagus (Asparagus officinalis L.): Changes in dietary fiber (nonstarch polysaccharides). J Agr Food Chem. 1999; 47: 3832–3836. [DOI] [PubMed] [Google Scholar]
- 53. Malm E, Bulone V, Wickholm K, Larsson PT, Iversen T. The surface structure of well-ordered native cellulose fibrils in contact with water. Carbohyd Res. 2010; 345: 97–100. 10.1016/j.carres.2009.10.020 [DOI] [PubMed] [Google Scholar]
- 54. Busse-Wicher M, Gomes TC, Tryfona T, Nikolovski N, Stott K, Grantham NJ, et al. The pattern of xylan acetylation suggests xylan may interact with cellulose microfibrils as a twofold helical screw in the secondary plant cell wall of Arabidopsis thaliana . Plant J. 2014; 79: 492–506. 10.1111/tpj.12575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Carpita NC, Gibeaut DM. Structural models of primary-cell walls in flowering plants—consistency of molecular-structure with the physical-properties of the walls during growth. Plant J. 1993; 3: 1–30. [DOI] [PubMed] [Google Scholar]
- 56. Yokoyama R, Nishitani K. Genomic basis for cell-wall diversity in plants. A comparative approach to gene families in rice and Arabidopsis. Plant Cell Physiol. 2004; 45: 1111–1121. [DOI] [PubMed] [Google Scholar]
- 57. Johnson MT, Carpenter EJ, Tian Z, Bruskiewich R, Burris JN, Carrigan CT, et al. Evaluating methods for isolating total RNA and predicting the success of sequencing phylogenetically diverse plant transcriptomes. Plos One. 2012; 7: e50226 10.1371/journal.pone.0050226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chiniquy D, Varanasi P, Oh T, Harholt J, Katnelson J, Singh S, et al. Three novel rice genes closely related to the Arabidopsis IRX9, IRX9L, and IRX14 genes and their roles in xylan biosynthesis. Front Plant Sci. 2013; 4: 83 10.3389/fpls.2013.00083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ren Y, Hansen SF, Ebert B, Lau J, Scheller HV. Site-directed mutagenesis of IRX9, IRX9L and IRX14 proteins involved in xylan biosynthesis: glycosyltransferase activity is not required for IRX9 function in Arabidopsis. Plos One. 2014; 9: e105014 10.1371/journal.pone.0105014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen XW, Vega-Sanchez ME, Verhertbruggen Y, Chiniquy D, Canlas PE, Fagerstrom A, et al. Inactivation of OsIRX10 leads to decreased xylan content in rice culm cell walls and improved biomass saccharification. Mol Plant. 2013; 6: 570–573. 10.1093/mp/sss135 [DOI] [PubMed] [Google Scholar]
- 61. Hornblad E, Ulfstedt M, Ronne H, Marchant A. Partial functional conservation of IRX10 homologs in physcomitrella patens and Arabidopsis thaliana indicates an evolutionary step contributing to vascular formation in land plants. BMC Plant Biol. 2013; 13: 3 10.1186/1471-2229-13-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kuhl JC, Cheung F, Yuan QP, Martin W, Zewdie Y, McCallum J, et al. A unique set of 11,008 onion expressed sequence tags reveals expressed sequence and genomic differences between the monocot orders Asparagales and Poales. Plant Cell. 2004; 16: 114–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kuhl JC, Havey MJ, Martin WJ, Cheung F, Yuan QP, Landherr L, et al. Comparative genomic analyses in Asparagus. Genome. 2005; 48: 1052–1060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Four predicted Asparagus sequences (AoIRX9, AoIRX9-L, AoIRX14_A and AoIRX14_B) were aligned with Arabidopsis GT43 proteins using Clustal X 2.1 and a neighbor-joining tree was produced. The bar represents a percent accepted mutations value of 10%. The numbers shown at branching points are bootstrap values derived from 1000 randomized samples.
(TIF)
IRX10 sequences were aligned using Clustal X 2.1. Alignment shown in Jalview 2.8 where blue shading indicates sequence identity across the five sequences. Greater than 80% identity is dark blue, greater than 60% is medium blue, greater than 40% is light blue and 40% or less has no shading.
(TIF)
Total RNA was extracted from apical, middle and basal sections of fresh Asparagus spears and the expression patterns of AoIRX9, AoIRX9-L, AoIRX10, AoIRX14_A, AoIRX14_B, PAL and NST1, were analyzed by semi-quantitative RT-PCR using actin as a control.
(TIF)
(DOC)
(DOC)
Data Availability Statement
The full-length sequences of these genes are available from the Genbank database (http://www.ncbi.nlm.nih.gov/Genbank) with the following accession numbers: AoIRX9 (KJ556998); AoIRX9-L (KJ556999); AoIRX10 (KJ557000); AoIRX14_A(KJ557001); AoIRX14_B (KJ557002).