Fig. 1.
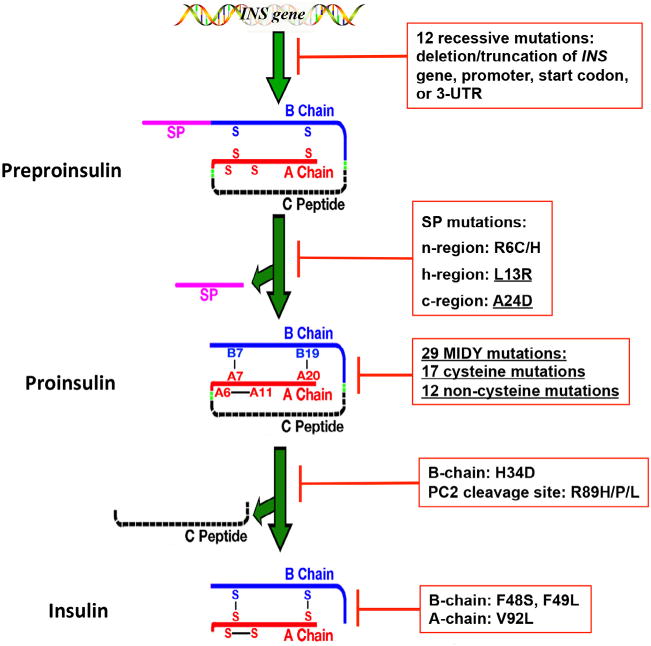
The effects of INS-gene mutations on the major steps of insulin biosynthesis. INS-gene mutations have been identified in the untranslated regions of INS gene and the coding sequence encoding all functional domains of preproinsulin molecule, including signal peptide (SP, pink), insulin B-chain (blue), C-peptide (black), insulin A-chain (red), and the proteolytic cleavage sites of signal peptidase (SPase) as well as prohormone convertases (PC1/3 and PC2, green). The mutations affect all major steps of insulin biosynthesis. Twelve recessive mutations in the untranslated regions result in more than 80% decrease of insulin production due to either INS gene deletion or truncation, or failure of insulin translation initiation, or instability of insulin mRNA. SP mutations in the n-region (R6C/H) or h-region (L13R) cause defective translocation of preproinsulin into the ER. The mutation at SP cleavage site (A24D) impairs SP cleavage. The largest group of INS-gene mutations are the mutations that affect proinsulin folding in the endoplasmic reticulum (ER), impairing formation of three evolutionarily conserved native disulfide bonds, B7-A7, B19-A20, and A6-A11. H34D affects sorting efficiency of proinsulin into regulated secretory pathway. The non-cysteine mutations at the PC2 cleavage site (R89H/P/L) impairs processing of proinsulin. The mutations in the B-chain (F48S and F49L) and A-chain (V92L) affect insulin binding to the insulin receptor. All mutations that cause Mutant INS-gene-induced Diabetes of Youth (MIDY) are underlined.
