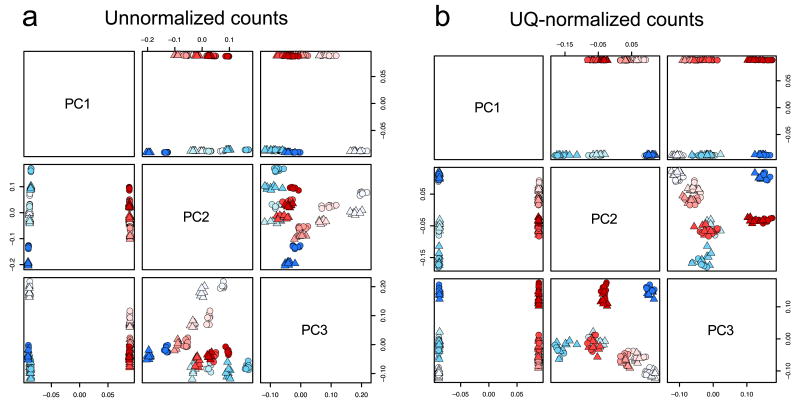
Figure 1.
Unwanted variation in S E Q C RNA-seq dataset. (a) Scatterplot matrix of first three principal components (PC) for unnormalized counts (log scale, centered). The PCs are orthogonal linear combinations of the original 21,559-dimensional gene expression profiles, with successively maximal variance across the 128 samples, i.e., the first PC is the weighted average of the 21,559 gene expression measures that provides the most separation between the 128 samples. Each point corresponds to one of the 128 samples. The four Sample A and the four Sample B libraries are represented by shades of blue and red, respectively (16 replicates per library). Circles and triangles represent samples sequenced in the first and second flow-cells, respectively. As expected for the SEQC dataset, the first PC is driven by the extreme biological difference between Sample A and Sample B. The second and third PCs clearly show library preparation effects (the samples cluster by shade) and, to a lesser extent, flow-cell effects reflecting differences in sequencing depths (within each shade, the samples cluster by shape). (b) Same as a, for upper quartile(UQ)–normalized counts. UQ normalization removes flow-cell effects (the circles and triangles now cluster together), but not library preparation effects. All other normalization procedures but RUV behave similarly as UQ (Supplementary Fig. 1).