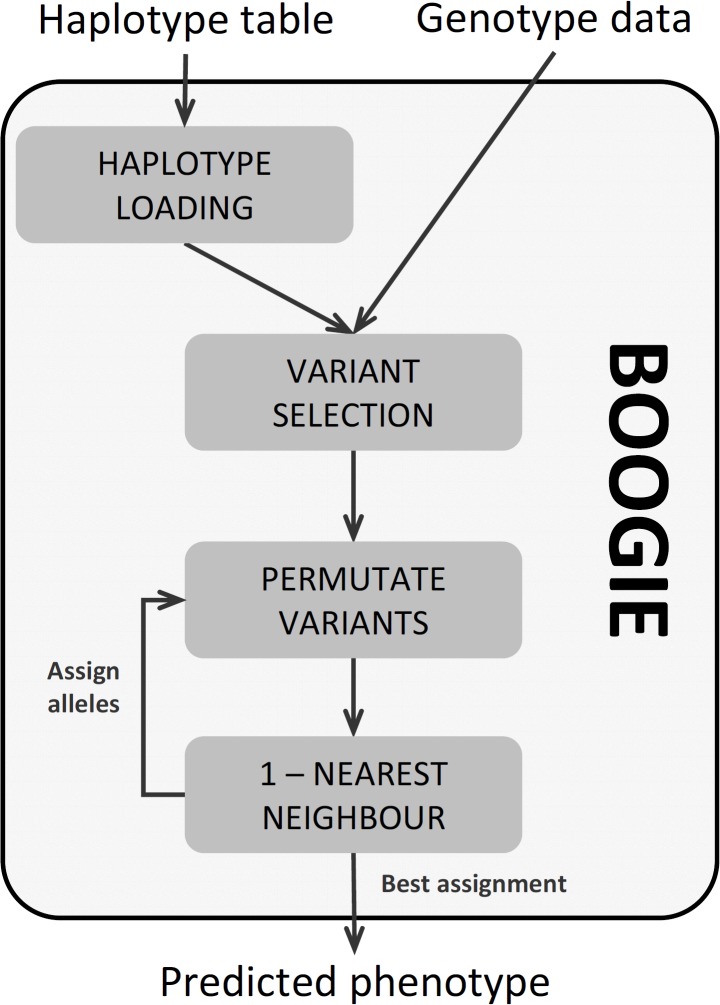
Fig 4. Schematic BOOGIE overview.
The tool requires just genotype data and an haplotype table for its execution. Genotype must be specified in a tabular file similar to VCF format, where chromosome, genomic position, nucleotides ad zygosity are specified. Haplotypes are defined in a tabular file, and each row specify the expected SNVs of a target phenotype. See README file of the application for format details. BOOGIE search for key variants in the input genotype, and optimize their assignment to a haplotypes with known phenotype according to the 1-nearest neighbour algorithm. The SNV permutation with best score is the one with highest phenotype likelihood.