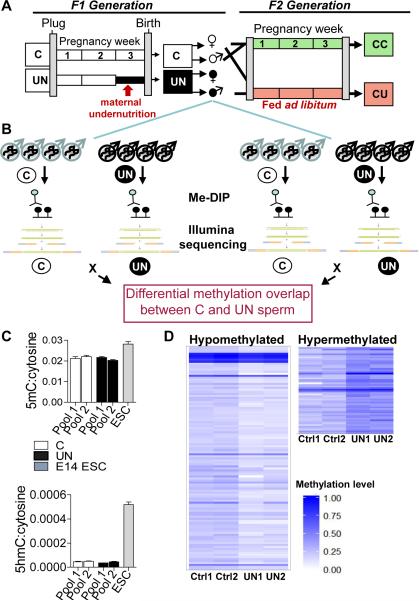
Figure 1. Total methylation is stable in UN sperm, with significant locus specific changes.
(A) Experimental design: F1 generation: Dams were randomised on pregnancy day 12.5 to control (C) or undernutrition (UN) groups and UN food intake restricted to 50%. Postnatal litters were equalised to eight pups and animals fed ad libitum. F2 generation: control F1 females mated at age 2 months with non-sibling control or UN males and fed ad libitum to produce: CC - both parents controls; CU - control dam, UN sire.
(B) Independent sperm DNA samples were quantified and pooled in equimolar ratios to generate two pools per condition. Control pools: n=8, 5 litters. UN pools: n=8, 4 litters. Following MeDIP-seq two independent C vs UN comparisons identified DMRs where methylation FC >1.5x and binomial p-value <0.0001 in both independent biological replicates.
(C) Mass spectrometry quantification of control and UN sperm 5-methyl-cytosine (above) and 5-hydroxymethyl-cytosine (below). E14 ESCs are shown for comparison.
(D) Heatmap of 111 hypomethylated DMRs (left) and 55 hypermethylated DMRs (right). Hypermethylated DMRs did not validate.