Figure 5.
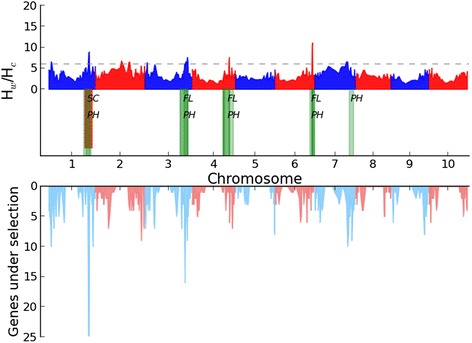
Genome-wide selection signatures. We calculated average ratios (expected heterozygosity of wild type sorghum (Hw) / expected heterozygosity of cultivated sorghum (Hc)) for windows of 500 consecutive SNPs throughout the genome. The gray dashed line is the cutoff for the top 5% of heterozygosity ratios. Green areas indicate confidence intervals determined by QTL mapping to contribute to either flowering time (FL) or plant height (PH), that also have strong selection signals. The hotspot on chromosome Sb01 identified by GWAS for pericarp color (SC) is highlighted by red. The distribution of 725 candidate genes implicated in domestication and/or improvement via gene-based population summary statistics [41] is shown below the heterozygosity ratios.
