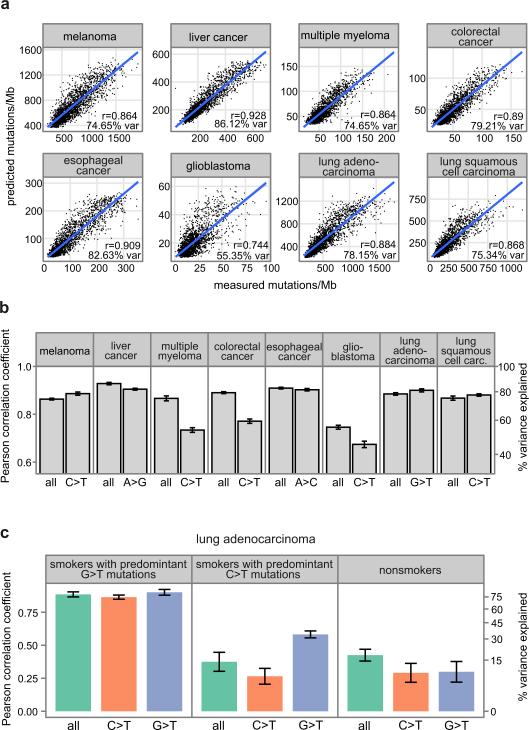
Figure 2.
Predicting local mutation density in cancer genomes using Random Forest regression trained on 424 epigenomic profiles. Pearson correlation between observed and predicted mutation densities along chromosomes is shown. (a) Actual versus predicted mutation densities in eight cancers. (b, c) Prediction accuracy represented as mean ± s.e.m (estimated using 10-fold cross-validation). Panels show prediction accuracy for all mutations and for nucleotide changes predominant in the corresponding cancer (b), and prediction accuracy in lung adenocarcinoma genomes stratified by smoking history and predominant nucleotide changes (G>T or C>T) (c).