Abstract
Protein kinase C (PKC) modulators are currently of great importance in preclinical and clinical studies directed at cancer, immunotherapy, HIV eradication, and Alzheimer’s disease. However, the bound conformation of PKC modulators in a membrane environment is not known. Rotational Echo Double Resonance (REDOR) NMR spectroscopy could uniquely address this challenge. However, REDOR NMR requires strategically-labeled, high affinity ligands to determine inter-label distances from which the conformation of the bound ligand in the PKC-ligand complex could be identified. Here we report the first computer-guided design and syntheses of three bryostatin analogs strategically-labeled for REDOR NMR analysis. Extensive computer analyses of energetically-accessible analog conformations suggested preferred labeling sites for the identification of the PKC-bound conformers. Significantly, three labeled analogs were synthesized, and, as required for REDOR analysis, all proved highly potent with PKC affinities (~1 nM) on par with bryostatin. These potent and strategically-labeled bryostatin analogs are new structural leads and provide the necessary starting point for projected efforts to determine the PKC-bound conformation of such analogs in a membrane environment, as needed to design new PKC modulators and understand PKC-ligand–membrane structure and dynamics.
INTRODUCTION
Identified by Nishizuka and co-workers in the 1970s,1 the PKC family of signaling proteins has since been implicated in a variety of normal and abnormal cellular processes associated with numerous therapeutic indications including cancer, immunotherapy, cardiovascular disease, HIV/AIDS eradication, and Alzheimer’s disease.2 PKC is regulated endogenously by mammalian diacyl glycerol (DAG). However, it is also activated by plant-derived phorbol esters (PEs).2a These agents (DAGs and PEs) compete for the same PKC binding site but upon binding, impart different biological functions. While DAG is required for normal signaling,3 PEs have been implicated in various other activities including tumor promotion. Bryostatin 1 (1, Figure 1, henceforth bryostatin), the first member of a novel marine macrolide family reported by Pettit and coworkers in 1982,4 was subsequently found to also compete with DAG and PEs in binding to PKC, but unlike the PEs, it is not a tumor promoter. Bryostatin shows efficacy against various cancer cell lines and has since been tested as a single agent and in combination therapy in several cancer clinical trials.5 More recently, bryostatin has been found to ameliorate ischemic damage6 and induce synaptogenesis,7 activities of potential clinical significance in the treatment of stroke and neurodegenerative diseases, respectively. The latter finding has led to its entry into a recently opened clinical trial for the treatment of Alzheimer’s disease.8 Bryostatin and its analogs have also been found to induce activation of latent HIV-infected cells,9,10 potentially leading to their elimination through a virus-induced cytopathic effect or host anti-HIV immunity. Agents that would eliminate such latent viral reservoirs, when used in conjunction with current antiretroviral therapy targeting the active virus, form the basis for a promising strategy to address an as yet unsolved problem, the eradication of HIV/AIDS.11
Figure 1.
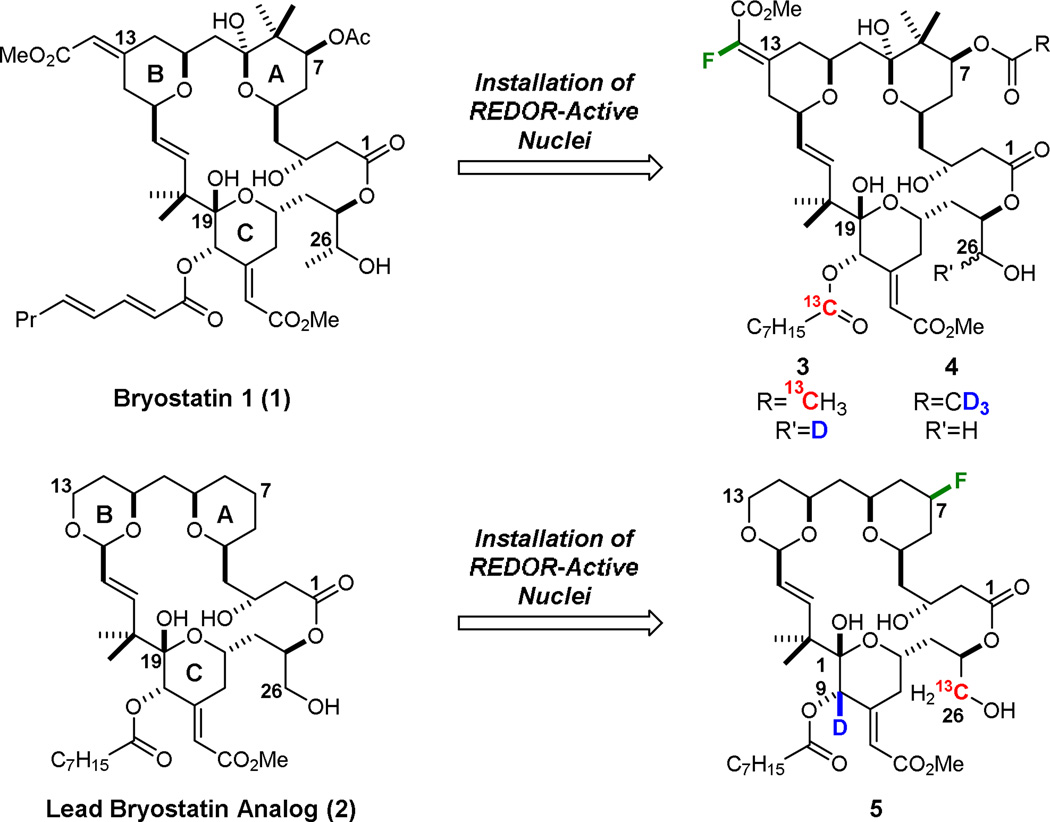
Labeled bryostatin analogs designed for REDOR NMR studies.
The diverse but highly significant activities associated with PKC modulators are partly a consequence of their differing selectivities for PKC isoforms.12 Three isoform classes have been identified: conventional PKCs: α, βI/βII, γ; novel PKCs: δ, ε, η, θ; and atypical PKCs. Conventional and novel PKCs are activated by DAG binding to PKC C1 domains, an interaction that requires PKC to transiently associate with a phospholipid bilayer where it phosphorylates various client proteins.13,14 Bryostatin, PEs and other natural scaffolds15 (e.g. aplysiatoxin and teleocidin) compete with endogenous DAG for binding to PKC but are orders of magnitude more potent than DAG and often elicit different biological activities. Based on a computational comparison of the binding elements in the preferred conformers of these naturally occurring agents,16 we reported the first designed PKC activators and subsequently used this pharmacophore analysis to design the first simplified but potent bryostatin analogs.17 This was followed by in silico docking efforts of these simplified analogs to the protein target.18 However, experimental support for these virtual structures is not available.
To understand how the PKC-ligand structure influences phenotypic function and thus how to design improved PKC modulators, accurate structural knowledge of the entire PKC-ligand-membrane complex is needed. Despite decades of effort focused on PKC and its C1 domains,15b this information does not yet exist.
Traditionally, X-ray crystallography or solution-state NMR have been employed to access structures of protein-ligand complexes, and these techniques have indeed contributed to our understanding of DAG- and bryostatin-responsive C1 domains.19 However, the vast majority of these efforts do not include any membrane-like component,20 a key feature given that that the ligand-PKC interaction and downstream phosphorylation events occur at the membrane.14 Indeed, our group has shown that the PKCγ C1b domain does not bind activators in buffered water but does bind the same activators upon addition of a membrane surrogate, specifically a vesicle composed of at least 15% phosphatidyl serine.21 These results have been recapitulated as part of two recent mutagenesis studies, demonstrating that both PKCδ22 and PKCθ19g C1b domains show diminished affinities for ligands when employing lipid mixtures of <20–30% phosphatidyl serine, thus reinforcing the importance of this membrane component for proper protein binding.
The only experimental efforts to date that interrogate a system with a lipid component rely on solution NMR studies of C1 domains in the presence of micelles, including those doped with diacylglycerols.23 However, these efforts have thus far focused on observing membrane surrogate-induced perturbations of the C1 domains, how these changes affect affinity for DAG, and how this data reinforces our current understanding of the step-by-step activation of PKC.24 The information obtained on the interactions between the ligand and the C1 domain (the area that would be the most useful for the design of preclinical candidates) while corroborating prior mutagenesis25 and other biochemical studies,19d,26 is pertinent only to the protein residues affected by DAG association. It does not provide information on the PKC bound ligand conformation. Molecular dynamics simulations have also been applied to these systems,23a,27 but again, the interaction between the membrane and the C1 domain, not structural detail on the bound ligand, was the primary focus. Given the potential clinical ramifications of controlling PKC, developing a more complete understanding of the structure and dynamics of the entire PKC-ligand-membrane complex is imperative to advancing current drug design efforts.
Toward this end, our efforts have focused on a strategy uniquely-suited for structural investigations of the protein-ligand-membrane systems, specifically Rotation Echo Double Resonance (REDOR) NMR spectroscopy. REDOR is a solid-state NMR technique that measures heteronuclear dipolar couplings between isolated pairs of NMR-active nuclei such as 2H, 13C, 15N, 19F, and 31P that can be translated into highly accurate inter-label distances of up to 16 Å between nuclei.28–30 Judicious positioning of labels within a ligand would allow inter-label distance determination, which upon computational analysis, would allow for identification of the conformation that best fit the bound ligand. As applied to bryostatin and the PKC system, intra-ligand, ligand-protein, intra-protein, ligand-membrane, and protein-membrane distances between suitably-labeled sites are possible, making this a uniquely effective approach to establishing the structure of complex protein-ligand-membrane systems. A critical prerequisite for a REDOR study is identifying label sites in a ligand that would allow for the translation of label distances into a unique bound structure. At the same time, the labeled ligand must be synthetically accessible, and for relevancy, it must exhibit high affinity binding.
Reported herein are the successful computer-guided design, synthesis, and biological evaluation of three new PKC modulatory ligands that meet the stringent criteria for REDOR analysis. Two new analogs (3 and 4) closely mimic natural bryostatins and the other (5) is analogous to one of the most potent and promising analogs reported to date by our group, B-ring dioxane analog 231 (a.k.a. picolog: Figure 1). While the affinities of these new, labeled analogs could not be predicted in advance, all three exhibit highly potent binding affinity (ca. 1 nM) to PKC C1 regulatory domains. These ligands thus successfully provide the required starting point for our projected studies directed at the first characterization of a PKC-ligand complex in a membrane-like microenvironment
RESULTS AND DISCUSSION
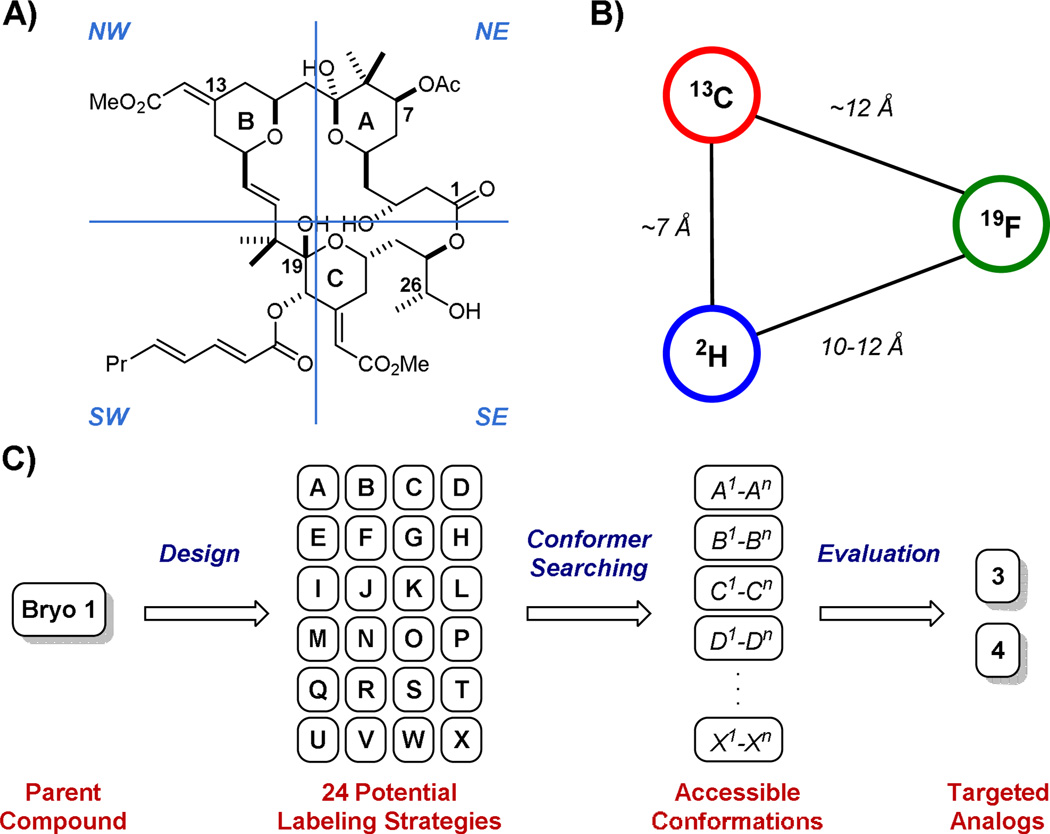
Bryostatin (1) and B-ring dioxane analog 2 provided reference points for the design of labeled analogs because the former is now in clinical trials and the latter represents one of our most studied and efficacious analogs, being as potent or more potent than bryostatin in growth inhibition assays involving 35 different human cancer cell lines in the National Cancer Institute screen and being well tolerated and more potent than bryostatin in an aggressive animal model of lymphoma.32 Our labeling strategy, a first of its kind for bryostatin, started with the recognition that bryostatin and analog 2 possess four spatially distinct domains or quadrants (Figure 2A) and that intra-domain mobility would be less than inter-domain mobility. Previous structure-function studies in our laboratory32,33 provided guidance on the selection of sites within each domain of this quadrant model that might tolerate a label without significantly reducing PKC affinity. However, adding to the risk and challenge of this project, affinities could only be determined after the compounds were made. While introduction of 2H- and 13C-labels carried little risk of affecting binding relative to unlabeled 1 and 2, the incorporation of the 19F-label incurred significant risk arising from potential electronic or hydrogen bonding effects. To ameliorate this potential problem, 19F-label placement was made distal to surfaces that are putatively in contact with the protein.
Figure 2.
Strategies for the Design of Bryostatin Analog Candidates for REDOR Analysis. A) Quadrant model applied to REDOR analog design; B) General distance constraints between nuclei in REDOR NMR; C) Graphical depiction of computer-guided analog selection process. Briefly, synthesis-informed design provided 24 potential labeling strategies (A–X), each representing a new compound and involving a 2H, 13C, and 19F labeling triad. Each structure could assume multiple (n) unique conformers as determined in silico where n ≥ 1 and can be different for each of A–X. Evaluation of labeling strategies as determined by their ability to distinguish between unique conformers left analogs 3 and 4 as the top targets. An analogous method was applied to B-ring dioxane analog 2 to select analog 5.
The dipolar couplings to be measured using REDOR are proportional to the product of the gyromagnetic ratios (γ) of the two nuclei of interest where γF > γC > γN > γD among commonly employed nuclei. Measuring couplings as small as 15 Hz is feasible with REDOR. Thus, for 13C-19F labeled pairs, inter-label distances up to 12 Å could be determined. 13C and 2H pairs require closer positioning in space as their coupling is comparatively weaker, setting the detection limit to ~7 Å. As such, 13C-19F or 2H-19F pairs feature prominently in the labeling array to permit longer range distance determinations (Figure 2B). Our labeling strategy also targeted core features of the scaffold, as the flexibility of side chains (e.g., the C20 octanoyl chain) would introduce a significantly larger number of available conformations and complicate interpretation of the REDOR analysis. Further, because inter-quadrant distances vary more than intra-quadrant distances for various conformers, labels were distributed amongst the four quadrants using a minimum of three labels for each ligand. The distances between these labels would ideally be different for each unique, energetically-accessible ligand conformation, allowing one to computationally differentiate between possible bound conformers based on experimentally-determined inter-label distances.
Guided by the above design criteria and by the need to identify synthetically-accessible ligands, specific labeling strategies arose iteratively from multiple in silico analyses of labeling options that would correspond to unique distances and thus structures (Figure 2C). First, a series of conformational searches were performed to sample the conformational space available to each of the two dozen bryostatin-like analogs that had been proposed as potential targets based on synthetic accessibility alone (the dioxane-based analog 5 was selected through an analogous method; further details are provided in the Supporting Information). Conformational searches were performed in a variety of solvents and force fields, generating from 4 to 26 unique conformational classes for each substrate. Importantly, the computational demand of this method was greatly reduced by using a stepwise approach to conformer generation.34 Given the numerous degrees of freedom in the C20 octanoyl side chain alone, conformers of only the macrocyclic core were initially evaluated. Upon elimination of redundant conformers, a side chain-only analysis followed by additional clustering provided an effectively comprehensive set of conformations accessible to each proposed labeled analog (see Figure 2C). The most effective labeling strategy was chosen based on in silico distances between labels, analyzing every conformational class amongst the potential analog library. Significantly, analogs 3, 4, and 5 each maintained at least two REDOR-accessible distances across all reasonable conformations. The combination of these distances is also expected to provide the most differentiation between conformational classes for any of the analogs considered, thus providing the best opportunity for obtaining a definitive bound conformer structure. As these labeled sites reside mostly on the edges of the molecule, they are also expected to be useful in future analyses targeting the interaction between the ligands and other components (e.g. key residues of the PKC C1 domains) in the solid-state sample.
For the bryostatin-like analogs 3 and 4, installation of the 19F label as an α-fluoro enoate ester at C13 was proposed, as it was predicted to be within range of the desired 13C-label at the carbonyl of the C20 octanoyl group (Figure 1) and potentially accessible through a previously unstudied olefination of the C13 ketone. Further, our group has previously observed PKC activity with both the E- and Z-enoate esters at C13,35 although the influence of the fluorine atom on both the stability and binding of such analogs was unknown. Two additional labels were to be included in analog 3, a 2-13C-acetoxy group at C7 (shift-resolved from the C20-octanoyl 13C label) and a deuterium at C26 which was to be installed as the final step from a C26 aldehyde. Analog 4 only incorporated one additional label, a C7 d3-acetoxy moiety, as the presence of three deuteriums is expected to greatly enhance the signal-tonoise for dipolar couplings to that position. Labeled Bring dioxane analog 5 incorporates an 19F label into the northeast quadrant (Figure 1), given the ability of that position to tolerate a variety of substituents without significant loss of affinity.36 To accommodate REDOR distance constraints while achieving potential conformer differentiation, siting a 13C label at C26 and 2H at C20 was proposed to complete labeling for simplified bryostatin analog 5.
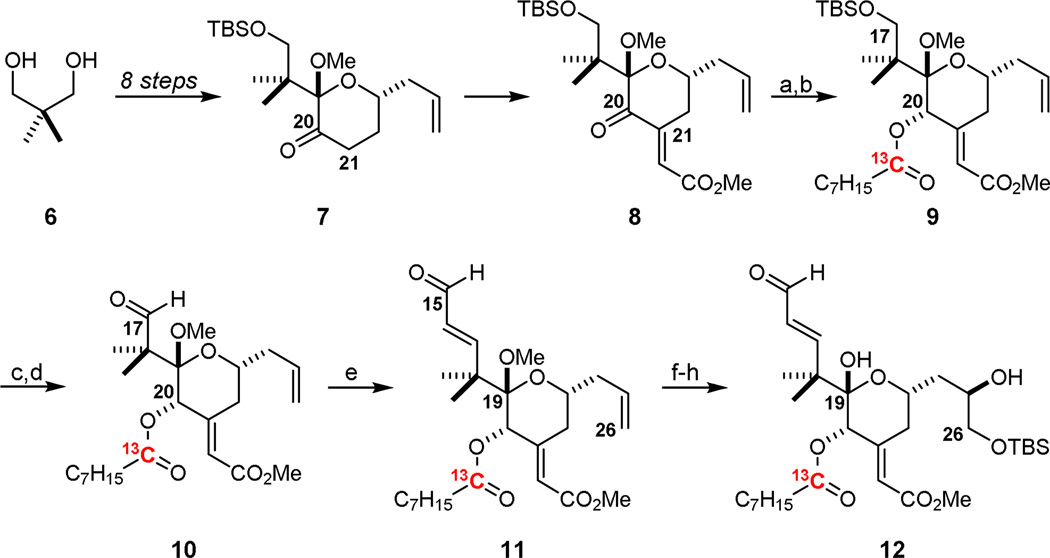
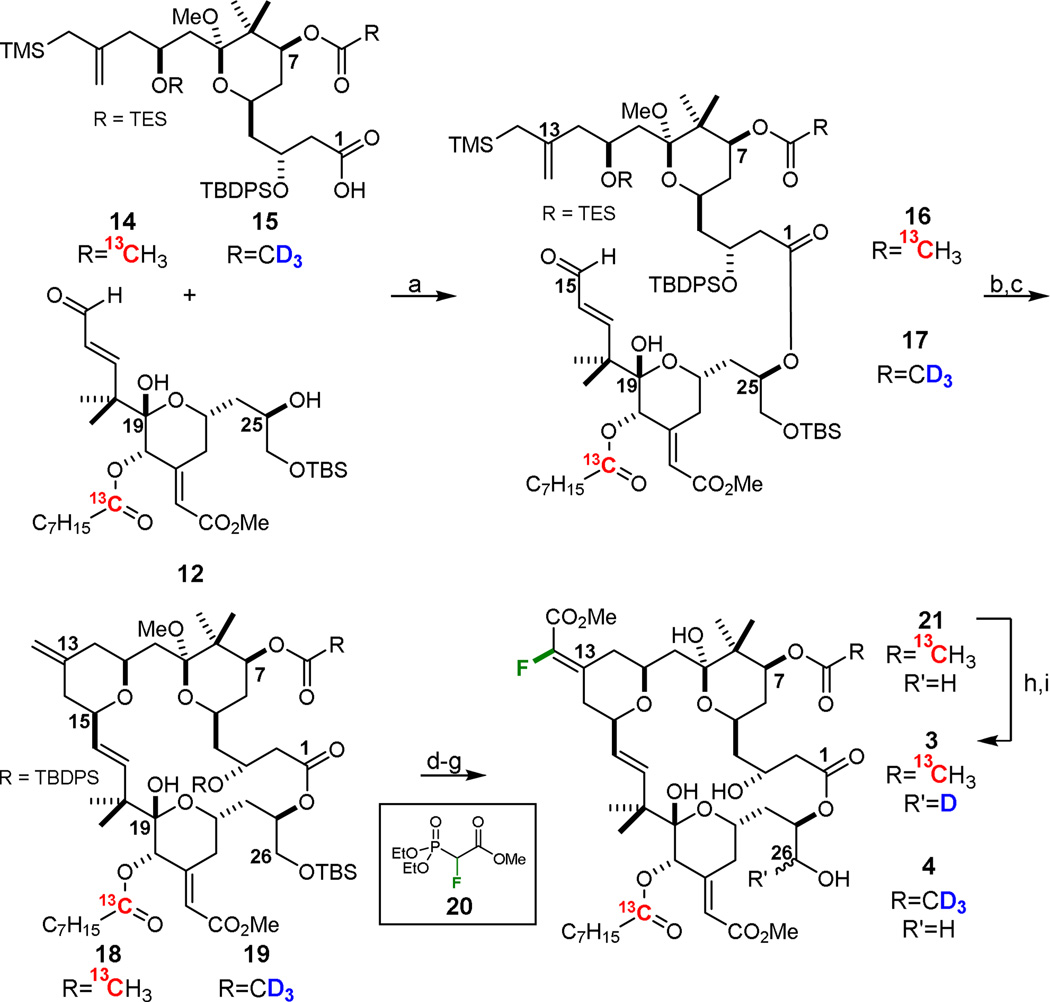
The convergent synthesis of the designed, labeled bryostatin analogs 3 and 4,37,38 which computationally met the stringent REDOR requirements, began with known intermediate 831a which is the aldol condensation product of methyl glyoxalate and ketone 7, a stable, storable intermediate prepared in multi-gram (>10 g) quantities in 8 steps from commercial materials (Scheme 1). Reduction with NaBH4 and DIC-mediated coupling with 1-13Coctanoic acid installed the desired 13C isotopic label in the C20 side chain. C17 silyl ether 9 was then deprotected with triethylamine trihydrogen fluoride, and subsequently oxidized to C17 aldehyde 10 with Dess-Martin periodinane. Homologation of 10 to the C15 enal 11 allowed for a Sharpless asymmetric dihydroxylation of the resultant C25-C26 olefin which proceeded to give a 2.7:1 ratio of diastereomers. After deprotection of the C19 hemiketal, selective TBS protection of the C26 primary alcohol afforded the desired 13C-labeled southern fragment 12 which was subsequently used to prepare both 3 and 4.
Scheme 1.
Synthesis of 13C-labeled southern fragment 12.a
aReagents and Conditions: a) NaBH4, CeCl3·7H2O, MeOH, −45 °C, dr > 20:1 α:β; b) 1-13C-octanoic acid, DIC, DMAP, CH2Cl2, rt, 76% over two steps; c) 3HF·NEt3, THF, rt; d) Dess-Martin periodinane, NaHCO3, CH2Cl2, rt, 73% over two steps; e) i. (Z)-2-bromo-1-ethoxyethylene, tBuLi, ZnMe2, ether, −78 °C, ii. 1 M aq. HCl, 80%; f) K2OsO2(OH)4, (DHQD)2PYR, K3Fe(CN)6, K2CO3, 1:1 tBuOH:H2O, 4 °C, 2.8:1 β:α dr; g) pTsOH·H2O, 4:1 MeCN:H2O, rt; h) TBSCl, imidazole, CH2Cl2, rt, 38% over three steps of pure C25 β-epimer.
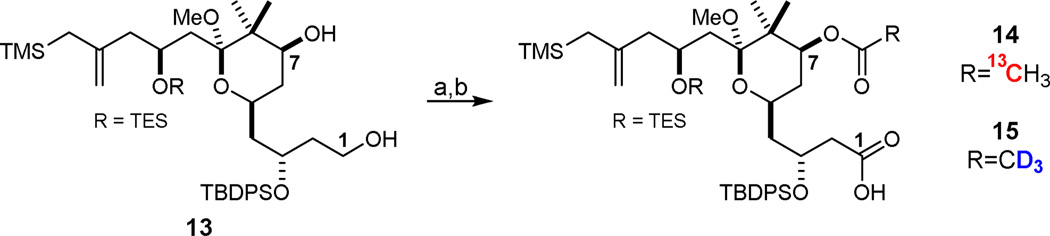
The northern fragments 14 and 15 were prepared in two steps from known diol 1337f using a one-flask TEMPO oxidation/ Pinnick oxidation procedure to yield the C1 carboxylic acid followed by acylation of C7 with 2,2’-13C-acetic anhydride or d6-acetic anhydride, respectively (Scheme 2).
Scheme 2.
Synthesis of northern fragments 14 and 15.a
aReagents and Conditions: a) i. TEMPO, PhI(OAc)2, 6:1 MeCN:H2O, rt, ii. NaClO2, NaH2PO4, 2-methyl-2-butene, H2O, 0 oC; For 14: b) 2,2’-13C-acetic anhydride, DMAP, CH2Cl2, −20 °C, 56% over two steps; For 15: b) d6-acetic anhydride, DMAP, CH2Cl2, −20 °C, 56% over two steps.
With the two fragments in hand for both analogs, the macrocycles were successfully formed through a two-step Yamaguchi esterification and Prins macrocyclization analogous to that employed in our synthesis of bryostatin 9 (Scheme 3).37f The resultant C13 olefins 18 and 19 were then each separately subjected to selective ozonolysis of the C13–C30 olefin, setting the stage for incorporation of a vinyl fluoride via a Horner-Wadsworth-Emmons protocol never before applied to this system. Importantly, the desired α-fluoro enoate ester was readily installed utilizing fluorinated phosphonoacetate 20.39 Interestingly, these HWE olefinations proceeded in relatively high diastereoselectivity (e.g. up to 5.5:1 toward 4) favoring the Z-enoate (equivalent to the E-enoate for the des-19F compound). This selectivity was unexpected given that olefination of analogous scaffolds under equivalent conditions with trimethyl phosphonoacetate proceed with little to no selectivity,37f,40,41 as would be expected for a ketone with only distal functionality differentiating the α and α’ carbons.42 However, these early HWE efforts were almost exclusively performed with NaHMDS as the base. A small screen of conditions (see Figure S6 in Supporting Information), revealed that n-BuLi favored higher diastereoselectivity (when using phosphonoacetate 20), perhaps suggesting a directing effect by the C9-OMe that requires the smaller Li counterion. Colder temperatures also bolstered selectivity, implying kinetic control.
Scheme 3.
End game strategy for analogs 3 and 4.a
aReagents and Conditions: For analog 3: a) i. 14, 2,4,6-trichlorobenzoyl chloride, NEt3, PhMe, rt, ii. 12, DMAP, PhMe, rt, 78%; b) PPTS, MeOH, rt; c) TBSCl, imidazole, CH2Cl2, rt, 57% over two steps; d) O3, CH2Cl2, MeOH, −78 °C; e) 20, NaHMDS, THF, 0 °C; f) HF·pyr, THF, rt; g) PPTS, 4:1 THF:H2O, rt; h) Dess-Martin periodinane, NaHCO3, CH2Cl2, rt; i) NaBD4, MeOH, −45 °C, 6% over six steps. For analog 4: a) i. 15, 2,4,6-trichlorobenzoyl chloride, NEt3, PhMe, rt, ii. 12, DMAP, PhMe, rt, 78%; b) PPTS, MeOH, rt; c) TBSCl, imidazole, CH2Cl2, rt, 65% over two steps; d) O3, CH2Cl2, MeOH, − 78 °C; e) 20, NaHMDS, THF, 0 °C, 5.5:1 z:E dr; f) HF·pyr, THF, rt; g) PPTS, 4:1 THF:H2O, rt, 7% over four steps.
Global deprotection with HF·pyridine produced the respective C9-α-fluoro ethers, which upon hydrolysis provided either des-2H intermediate 21 or completed analog 4. The deuterium label was installed through Dess-Martin periodinane mediated oxidation of 21 at C26, and subsequent reduction with NaBD4, to give bryostatin analog 3 as a 1:1 mixture of C26 epimers.
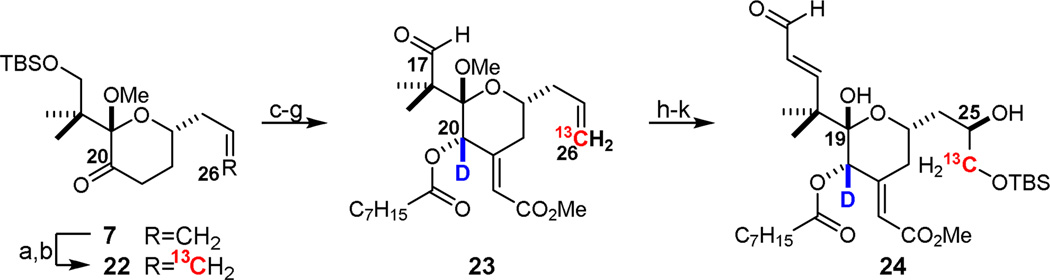
Synthesis of the southern fragment of analog 5 followed closely the route used above to prepare 12. To install the desired 13C label at C26, ketone 7 was ozonolyzed and the product converted by a Wittig olefination to labeled ketone 22 (Scheme 4). A synthetic sequence involving aldol condensation, reduction of C20 with NaBD4, acylation with octanoic anhydride, deprotection of the C17 silyl ether, and oxidation at C17 afforded the bis-labeled aldehyde intermediate 23. Homologation to the C15 enal, asymmetric dihydroxylation of the C25-C26 alkene, deprotection of C19, and protection of the newly formed C26 alcohol were performed as described above, providing 2H,13C-labeled southern fragment 24.
Scheme 4.
Synthesis of bis-labeled southern fragment 24.a
aReagents and Conditions: a) i. O3, CH2Cl2, −78 °C, ii. PPh3, rt; b) Ph3P13CH3I, KHMDS, PhMe, −78 °C, 74% over two steps; c) methyl glyoxalate, K2CO3, MeOH, rt, 78%; d) NaBD4, CeCl3·7H2O, MeOH, −45 °C, dr > 20:1 α:β; e) octanoic anhydride, DMAP, CH2Cl2, rt, 83% over two steps; f) 3HF·NEt3, THF, rt; g) Dess-Martin periodinane, NaHCO3, CH2Cl2, rt; h) i. (Z)-2-bromo-1-ethoxyethylene, tBuLi, ZnMe2, ether, −78 °C, ii. 1 M aq. HCl, 45% over three steps; i) K2OsO2(OH)4, (DHQD)2PYR, K3Fe(CN)6, K2CO3, 1:1 tBuOH:H2O, 4 °C, 2.5:1 β:α dr; j) pTsOH·H2O, 4:1 MeCN:H2O, rt; k) TBSCl, imidazole, CH2Cl2, rt, 39% over three steps of pure C25 β-epimer.
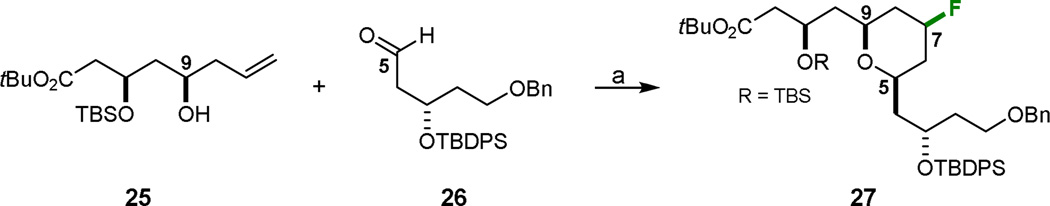
Synthesis of the northern fragment 32 began with a facile, convergent approach to the preparation of A-ring tetrahydropyran 27 (Scheme 5). Taking inspiration from the fluoride-trapped Prins macrocyclization recently reported by our group,43 aldehyde 2644 and homoallylic alcohol 25 were coupled with concomitant fluoride incorporation using BF3·OEt2. However, low yields (~15%) and difficulties associated with separation from various diastereomers and byproducts (competing elimination pathways) precluded advancement of this strategy. Intramolecular variants of this closure (based on the work of Rycknovsky45) as well as changing fluoride sources (e.g. DAST, anhydrous TBAF, HF·pyr, 3HF·NEt3) did not improve the outcome.
Scheme 5.
Initial synthesis of C7 fluoride 27 via fluoro-Prins cyclization.a
aReagents and Conditions: a) BF3·OEt2, ether, −45 °C to rt, ~15%.
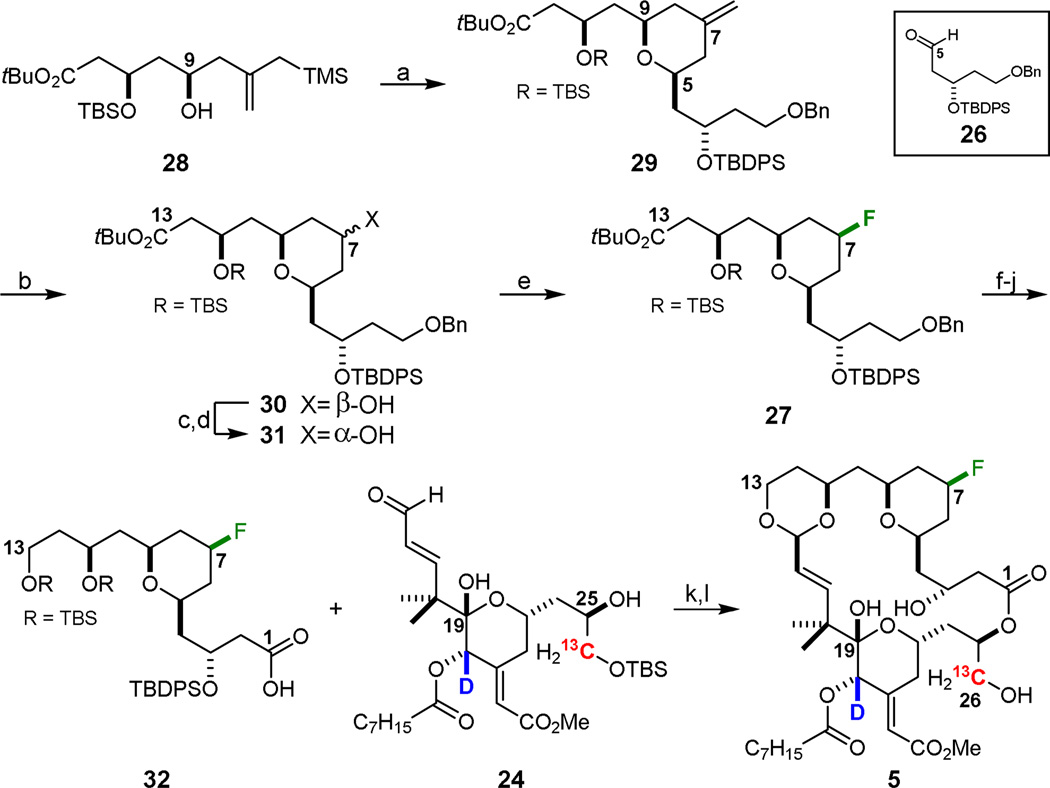
A more efficient synthesis of northern fragment (32) of analog 5 was ultimately realized using a silyl-terminated Prins closure sequence (Scheme 6). Northern fragment 32 was prepared from previously reported homoallylic alcohol 2836 Prins cyclization with aldehyde 26 cleanly afforded pyran 29 in 89% yield. Subsequent ozonolysis gave the C7 ketone and reduction with sodium borohydride selectively generated the C7 β-alcohol 30 in near quantitative yield. Inversion of stereochemistry through a Mitsunobu/methanolysis sequence provided C7 α-alcohol 31, setting the stage for the 19F installation. DAST-mediated deoxyfluorination was then used to generate C7 fluoride 27. Elimination byproducts (C6–C7 and C7–C8 olefins) accounted for 15–20% of the yield loss. Reduction and TBS protection of C13 afforded the corresponding tris-silyl ether after which the C1 benzyl ether was removed. Finally, oxidation of the resulting primary alcohol yielded the required C1 carboxylic acid of northern fragment 32. The isotopically-labeled dioxane analog 5 was then obtained through Yamaguchi esterification of C25 on southern fragment 24 with northern fragment 32, followed by a remarkable one-step global deprotection and macroacetalization that formed the B-ring, set the C15 stereocenter, and closed the macrocycle.
Scheme 6.
Synthesis of 19F-labeled northern fragment 32 and elaboration to REDOR analog 5.a
aReagents and Conditions: a) TMSOTf, ether, −78 °C, 89%; b) i. O3, 6:1 CH2Cl2:MeOH, −78 °C, ii. NaBH4, −78 °C to rt, 98%; c) p-NO2-benzoic acid, DIAD, PPh3, PhMe, rt; d) K2CO3, MeOH, rt, 75% over two steps; e) DAST, CH2Cl2, −78 °C, 50%; f) LiBH4, THF, 35 °C; g) TBSCl, imidazole, CH2Cl2, rt, 78% over two steps; h) lithium naphthalenide, THF, −25 °C; i) TPAP, NMO, CH2Cl2, 4A MS, rt; j) NaClO2, NaH2PO4, 2-methyl-2-butene, 2:1 tBuOH:H2O, 0 °C, 74% over three steps; k) i. 32, 2,4,6-trichlorobenzoyl chloride, NEt3, PhMe, rt, ii. 24, DMAP, PhMe, rt; l) HF·pyr, THF, rt, 56% over two steps.
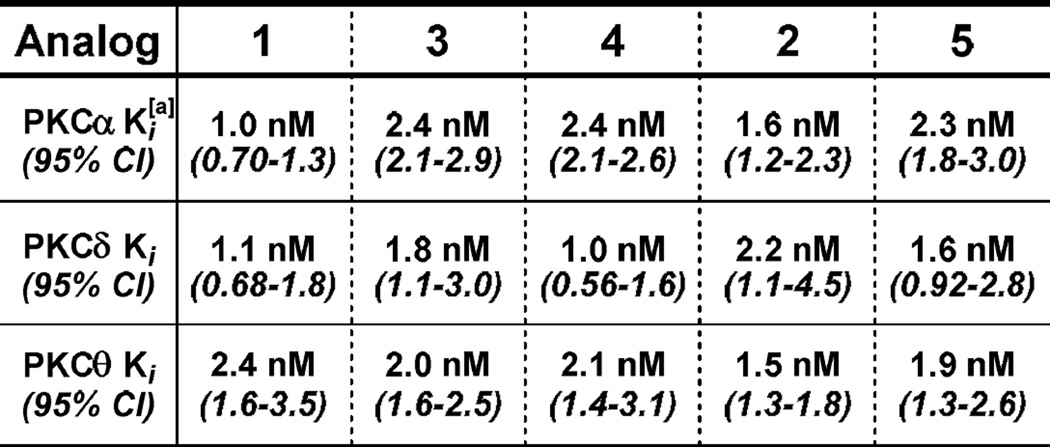
The stage was thus set for a crucial test, i.e. determining whether these analogs would bind PKC with sufficiently high affinity to be suitable candidates for REDOR NMR experiments. The REDOR measurements would be performed on biologically relevant substrate-ligand complexes in a membrane microenvironment, and the total amount of ligand in the final sample thus depends on its binding affinity and concentration. High affinity ligands (similar to the parent bryostatin) would maximize the amount of bound ligand and thus maximize the sensitivity of the measurements, an important consideration given the anticipated long acquisition times (from days to weeks or even up to months) required to accurately measure the desired long-range distances. Significantly, all three isotopically-labeled analogs (3, 4, and 5) exhibited PKC affinities comparable to bryostatin and B-ring dioxane analog 2 across three separate isoforms: α, δ, and θ (see Figure 3). Thus, these compounds not only meet the stringent requirements for a REDOR study, but also represent entirely new leads for ongoing structure-function relationships of PKC modulators, now with novel fluorine installations. The isoforms chosen have all been studied from a structural perspective before, thereby allowing for direct comparisons between experimental techniques once REDOR data is obtained. With these potent analogs in hand, the first-in-class interrogation of a PKC-ligand complex in a membrane environment can now be approached using REDOR NMR. These REDOR NMR analyses will be reported in due course. These studies are expected to enhance drug design efforts targeting this central signaling pathway, potentially leading to more effective pre-clinical leads and more generally to a greater understanding of protein-ligand binding and function in a membrane microenvironment. Notably, the experimentally-obtained affinities validate the computation- and synthesis-informed design strategy detailed above, suggesting the potential for application to other flexible, bioactive scaffolds that would benefit from REDOR analysis.
Figure 3.
Ki Values of Isotopically-Labeled Bryostatin Analogs Against PKC Isoforms.[a] Values obtained from heterogeneous competitive binding assay with [3H]-phorbol diacetate46; 95% CI = 95% confidence interval.
CONCLUSION
The PKC family of signaling proteins is implicated in normal cellular function and in diseases for which few or no therapeutic solutions exist. The bound conformation of ligands that regulate PKC isoforms in a membrane microenvironment is not known but required to understand ligand function and inform efforts toward isoform selective modulation. The studies reported herein demonstrate the successful development of bryostatin-based molecular probes designed to determine intra-nuclear distances of the bound probe and thus the structure of ligand-PKC complexes in a membrane microenvironment. A comprehensive computational analysis has been used to develop a strategy for labeling bryostatin analogs for REDOR experiments. This strategy led to the design and successful scalable synthesis of three new, potent ligands and candidates for REDOR NMR analysis. These highly convergent syntheses show the generality of our approaches to the synthesis of bryostatin analogs including key macroacetalization and Prins macrocyclization ring closure strategies, while also demonstrating techniques for the introduction of NMR-active nuclei into the scaffold. Extending beyond what could be predicted at the outset, the three designed analogs showed potent PKC affinities on par with bryostatin itself. This study thus sets the stage for first-in-class REDOR NMR experiments designed to access structural information on the analog-PKC complex in a membrane microenvironment as required for the design of new analogs of current and projected clinical interest related to cancer, Alzheimer’s disease, and HIV/AIDS eradication.
Supplementary Material
ACKNOWLEDGMENT
This research was supported by the National Institutes of Health (CA031845 and CA031841 to P.W.; DP2 OD007488 to L.C.). Additional funding was provided by the Stanford University Center for Molecular Analysis and Design (B.L.) and the Amgen Graduate Fellowship (D.S.).
Footnotes
ASSOCIATED CONTENT
Description of computational methods, including conformational search parameters. Synthetic procedures, characterization data, and 1H and 13C spectra for all novel compounds. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.a) Inoue M, Kishimoto A, Takai Y, Nishizuka Y. J. Biol. Chem. 1977;252:7610–7616. [PubMed] [Google Scholar]; b) Nakamura S, Yamamura H. J. Biochem. 2010;148:125–130. doi: 10.1093/jb/mvq066. [DOI] [PubMed] [Google Scholar]
- 2.a) Nishizuka Y. Nature. 1984;308:693–698. doi: 10.1038/308693a0. [DOI] [PubMed] [Google Scholar]; b) Battaini F, Mochly-Rosen D. Pharmacol. Res. 2007;55:461–466. doi: 10.1016/j.phrs.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Mochly-Rosen D, Das K, Grimes K. Nature Rev. Drug Disc. 2012;11:937–957. doi: 10.1038/nrd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newton A. Am. J. Physiol. Endocrinol. Metab. 2010;298:E395–E402. doi: 10.1152/ajpendo.00477.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pettit G, Herald C, Doubek D, Herald D, Arnold E, Clardy J. J. Am. Chem. Soc. 1982;104:6846–6848. [Google Scholar]
- 5.a) For current clinical information, see: http://clinicaltrials.gov; Barr PM, Lazarus HM, Cooper BW, Schluchter MD, Panneerselvam A, Jacobberger JW, Hsu JW, Janakiraman N, Simic A, Dowlati A, Remick SC. Am. J. Hematol. 2009;84:484. doi: 10.1002/ajh.21449. Kortmansky J, Schartz G. Cancer Invest. 2003;21:924–936. doi: 10.1081/cnv-120025095. Gennas G, Talman V, Yli-Kauhaluoma J, Tuominen R, Ekokoski E. Curr. Top. Med Chem. 2011;11:1370–1392. doi: 10.2174/156802611795589584.
- 6.a) Sun M, Hongpaisan J, Nelson T, Alkon D. Proc. Natl. Acad. Sci. USA. 2008;105:13620–13625. doi: 10.1073/pnas.0805952105. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zohar O, Lavy R, Zi X, Nelson T, Hongpaisan J, Pick C, Alkon D. Neurobiol. Dis. 2011;41:329–337. doi: 10.1016/j.nbd.2010.10.001. [DOI] [PubMed] [Google Scholar]; c) Tan Z, Turner R, Leon R, Li X, Hongpaisan J, Zheng W, Logsdon A, Naser Z, Alkon D, Rosen C, Huber J. Stroke. 2013;44:3490–3497. doi: 10.1161/STROKEAHA.113.002411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Hongpaisan J, Alkon D. Proc. Natl. Acad. Sci. USA. 2007;104:19571–19576. doi: 10.1073/pnas.0709311104. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sun M, Hongpaisan J, Alkon D. Proc. Natl. Acad. Sci. USA. 2009;106:14676–14680. doi: 10.1073/pnas.0907842106. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Khan T, Nelson T, Verma V, Wender P, Alkon D. Neurobiol. Dis. 2009;34:332–339. doi: 10.1016/j.nbd.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Sun M, Alkon D. Pharmacol. Therapeut. 2010;127:66–77. doi: 10.1016/j.pharmthera.2010.03.001. [DOI] [PubMed] [Google Scholar]; e) Kim H, Han S, Quan H, Jung Y, An J, Kang P, Park J, Yoon B, Seol G, Min S. Neuroscience. 2012;226:348–355. doi: 10.1016/j.neuroscience.2012.08.055. [DOI] [PubMed] [Google Scholar]
- 8.Trial is still recruiting as of Jan. 14th, 2015 (NCT02221947).
- 9.Mehla R, Bivalkar-Mehla S, Zhang R, Handy I, Albrecht H, Giri S, Nagarkatti P, Nagarkatti M, Chauhan A. PLoS One. 2010;5:e11160. doi: 10.1371/journal.pone.0011160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeChristopher B, Loy B, Marsden M, Schrier A, Zack J, Wender P. Nature Chem. 2012;4:705–710. doi: 10.1038/nchem.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.For the current status of latency-reversing agents in HIV therapy research along with challenges of this treatment method see: Chan CN, Dietrich I, Hosie M, Willet B. J. Gen. Virol. 2013;94:917–932. doi: 10.1099/vir.0.049296-0. Bullen C, Laird G, Durand C, Siliciano J, Siliciano R. Nature Med. 2014;20:425–430. doi: 10.1038/nm.3489. Siliciano J, Siliciano R. J. Allergy Clin Immunol. 2014;134:12–19. doi: 10.1016/j.jaci.2014.05.026. Barouch D, Deeks S. Science. 2014;345:169–174. doi: 10.1126/science.1255512. Archin N, Marsh Sung J, Garrido C, Soriano-Sarabia N, Margolis D. Nat. Rev. Microbiol. 2014;12:750–764. doi: 10.1038/nrmicro3352.
- 12.a) See reference 2c; b) Several other C1-domain containing proteins are possible targets for bryostatin see: Kazanietz MG. Mol. Pharmacol. 2002;61:759–767. doi: 10.1124/mol.61.4.759.
- 13.For reviews of PKC see: Steinberg S. Physiol. Rev. 2008;88:1341–1378. doi: 10.1152/physrev.00034.2007. Wu-Zhang A, Newton A. Biochem. J. 2013;452:195–209. doi: 10.1042/BJ20130220. as well as references 2c and 3.
- 14.For recent literature investigating the nuances of PKC activation see: Ziemba B, Li J, Landgraf K, Knight J, Voth G, Falke J. Biochemistry. 2014;53:1697–1713. doi: 10.1021/bi4016082. Antal C, Violin J, Kunkel M, Skovso S, Newton A. Chem. Biol. 2014;21:459–469. doi: 10.1016/j.chembiol.2014.02.008.
- 15.a) Irie K, Yanagita R. Chem. Rec. 2014;14:251–267. doi: 10.1002/tcr.201300036. [DOI] [PubMed] [Google Scholar]; b) Das J, Rahman G. Chem. Rev. 2014;114:12108–12131. doi: 10.1021/cr300481j. [DOI] [PubMed] [Google Scholar]
- 16.Wender P, Koehler K, Sharkey N, Dell’Aquila M, Blumberg P. Proc. Natl. Acad. Sci. USA. 1986;83:4214–4218. doi: 10.1073/pnas.83.12.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wender P, Cribbs C, Koehler K, Sharkey N, Herald C, Kamano Y, Pettit G, Blumberg P. Proc. Natl. Acad. Sci. USA. 1988;85:7197–7201. doi: 10.1073/pnas.85.19.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wender P, DeBrabander J, Harran P, Jimenez J-M, Koehler M, Lippa B, Park C-M, Siedenbiedel C, Pettit G. Proc. Natl. Acad. Sci. USA. 1998;95:6624–6629. doi: 10.1073/pnas.95.12.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.a) Wender P, Baryza J, Brenner S, Clark M, Craske M, Horan J, Meyer T. Curr. Drug. Dis. Tech. 2004;1:1–11. doi: 10.2174/1570163043484888. [DOI] [PubMed] [Google Scholar]; b) Keck G, Poudel Y, Rudra A, Stephens J, Kedei N, Lewin N, Peach M, Blumberg P. Angew. Chem. Int Ed. 2010;94:4580–4584. doi: 10.1002/anie.201001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.a) Hommel U, Zurini M, Luyten M. Struct. Bio. 1994;1:383–387. doi: 10.1038/nsb0694-383. [DOI] [PubMed] [Google Scholar]; b) Xu R, Pawelczyk T, Xia TH, Brown S. Biochem. 1997;36:10709–10717. doi: 10.1021/bi970833a. [DOI] [PubMed] [Google Scholar]; c) Ichikawa S, Hatanaka H, Takeuchi Y, Ohno S, Inagaki F. J. Biochem. 1995;117:566–574. doi: 10.1093/oxfordjournals.jbchem.a124745. [DOI] [PubMed] [Google Scholar]; d) Zhang G, Kazanietz M, Blumberg P, Hurley J. Cell. 1995;81:917–924. doi: 10.1016/0092-8674(95)90011-x. [DOI] [PubMed] [Google Scholar]; e) Leonard T, Rozycki B, Saidi L, Hummer G, Hurley J. Cell. 2011;144:55–66. doi: 10.1016/j.cell.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Shanmugasundararaj S, Das, Sandberg W, Zhou X, Wang D, Messing R, Bruzik K, Stehle T, Miller K. Biophys. J. 2012;103:2331–2340. doi: 10.1016/j.bpj.2012.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Rahman G, Shanker S, Lewin N, Kedei N, Hill C, Prasad V, Blumberg P, Das J. Biochem. J. 2013;451:33–44. doi: 10.1042/BJ20121307. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Ziemba B, Booth J, Jones D. Biomol. NMR Assign. 2011;5:125–129. doi: 10.1007/s12104-010-9283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Notably, computational analysis of the binding pockets revealed by these various structural studies showed no correlation between volume available and affinity for ligands suggesting a disconnect between current structural data and observed biological activity: Rahman G, Das J. J. Biomol. Struct. Dyn. 2015;33:219–232. doi: 10.1080/07391102.2014.895679.
- 21.Wender P, Irie K, Miller B. Proc. Natl. Acad. Sci. USA. 1995;92:239–243. doi: 10.1073/pnas.92.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelsey J, Geczy T, Lewin N, Kedei N, Hill C, Selezneva J, Valle C, Woo W, Gorshkova I, Blumberg P. ChemBioChem. 2014;15:1131–1144. doi: 10.1002/cbic.201400041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.a) Stewart M, Morgan B, Massi F, Igumenova T. J. Mol. Biol. 2011;408:949–970. doi: 10.1016/j.jmb.2011.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Stewart M, Cole T, Igumenova T. J. Biol. Chem. 2014;289:27653–27664. doi: 10.1074/jbc.M114.585570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phorbol dibutyrate was also used as a reference ligand, though the endogenous ligand was the clear focus of both studies.
- 25.Kazanietz M, Wang S, Milne G, Lewin N, Liu H, Blumberg P. J. Biol. Chem. 1995;270:21852–21859. doi: 10.1074/jbc.270.37.21852. [DOI] [PubMed] [Google Scholar]
- 26.For example: Dries D, Gallegos L, Newton A. J. Biol. Chem. 2007;282:826–830. doi: 10.1074/jbc.C600268200.
- 27.Li J, Ziemba B, Falke J, Voth G. J. Am. Chem. Soc. 2014;136:11757–11766. doi: 10.1021/ja505369r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seminal contribution: Gullion T, Schaefer J. J. Magn. Reson. 1989;81:196–200. For pertinent reviews see: Toke O, Cegelski L. In: Encyclopedia of Magnetic Resonance. Harris R, Wasylishen R, editors. Weinheim: WILEY-VCH; 2010. Matsuoka S, Inoue M. Chem. Commun. 2009:5664–5675. doi: 10.1039/b910230b. Cegelski L. Bioorg. Med. Chem. Lett. 2013;23:5767–5775. doi: 10.1016/j.bmcl.2013.08.064.
- 29.Louie E, Chirakul P, Raghunathan V, Sigurdsson S, Drobny G. J. Magn. Reson. 2006;178:11–24. doi: 10.1016/j.jmr.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 30.Paik Y, Yang C, Metaferia B, Tang S, Bane S, Ravindra R, Shanker N, Alcaraz A, Johnson S, Schaefer J, O’Connor R, Cegelski L, Snyder J, Kingston D. J. Am. Chem. Soc. 2007;129:361–370. doi: 10.1021/ja0656604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.a) Wender P, Baryza J, Bennett C, Bi C, Brenner S, Clarke M, Horan J, Kan C, Lacote E, Lippa B, Nell P, Turner T. J. Am. Chem. Soc. 2002;124:13648–13649. doi: 10.1021/ja027509+. [DOI] [PubMed] [Google Scholar]; b) DeChristopher B, Fan A, Felsher D, Wender P. Oncotarget. 2012;3:58–66. doi: 10.18632/oncotarget.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wender P, Donnelly A, Loy B, Near K, Staveness D. In: Natural Products in Medicinal Chemistry. Hanessian S, editor. Weinheim: Wiley-VCH Verlag GmbH & Co. KGaA; 2014. pp. 475–544. [Google Scholar]
- 33.Other labs have also contributed analog strategies, with the most recent efforts being shown here: Kraft M, Poudel Y, Kedei N, Lewin N, Peach M, Blumberg P, Keck G. J. Am. Chem. Soc. 2014;136:13202–13208. doi: 10.1021/ja5078188. Andrews I, Ketcham J, Blumberg P, Kedei N, Lewin N, Peach M, Krische M. J. Am. Chem. Soc. 2014;136:13209–13216. doi: 10.1021/ja507825s.
- 34.Kimura K, Mizutani M, Tomioka N, Endo Y, Shudo K, Itai A. Chem. Pharm. Bull. (Tokyo) 1999;47:1134–1137. [Google Scholar]
- 35.Wender P, DeChristopher B, Schrier A. J. Am. Chem. Soc. 2008;130:6658–6659. doi: 10.1021/ja8015632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.While hydrogen bond donors at C7 abrogate activity, most other polar functionality is tolerated: Wender P, Verma V. Org. Lett. 2008;10:3331–3334. doi: 10.1021/ol801235h.
- 37.Decades of synthetic effort have been invested in the synthesis of bryostatins. Seven total syntheses have been realized in that time: Kageyama M, Tamura T, Nantz MH, Roberts JC, Somfai P, Whritenour DC, Masamune S. J. Am. Chem. Soc. 1990;112:7407–7408. Evans DA, Carter PH, Carreira EM, Charette AB, Prunet JA, Lautens M. J. Am. Chem. Soc. 1999;121:7540–7552. Ohmori K, Ogawa Y, Obitsu T, Ishikawa Y, Nishiyama S, Yamamura S. Angew. Chem. Int. Ed. 2000;39:2290–2294. Trost BM, Dong G. Nature. 2008;456:485–488. doi: 10.1038/nature07543. Keck GE, Poudel YB, Cummins TJ, Rudra A, Covel JA. J. Am. Chem. Soc. 2011;133:744–747. doi: 10.1021/ja110198y. Wender PA, Schrier AJ. J. Am. Chem. Soc. 2011;133:9228–9231. doi: 10.1021/ja203034k. Lu Y, Woo SK, Krische MJ. J. Am. Chem. Soc. 2011;133:13876–13879. doi: 10.1021/ja205673e. h) a formal total synthesis of bryostatin 7 has also been accomplished: Manaviazar S, Frigerio M, Bhatia GS, Hummersone MG, Aliev AE, Hale KJ. Org. Lett. 2006;8:476–4480. doi: 10.1021/ol061626i. i) Ref. 32 provides some detail on each of the above total syntheses.
- 38.Several other groups have provided partial syntheses, these are reviewed here: a) 1982–2000: Hale KJ, Hummersone MG, Manaviazar S, Frigerio M. Nat. Prod. Rep. 2002;19:413–453. doi: 10.1039/b009211h. b) 2001–2009: Hale KJ, Manaviazar S. Chem. Asian J. 2010;5:704–754. doi: 10.1002/asia.200900634.
- 39.Prepared in one step from commercial materials see: Remme N, Koschek K, Schneider C. Synlett. 2007;3:491–493.
- 40.Minor selectivities have previously been observed in HWE olefinations of C13 ketone intermediates but not to the extent seen here (note: the Z-isomer of analogs 3 and 4 is equivalent to the E-isomer of des-fluoro species). These examples can be found here: a) 1:1.6 E:Z in Ohmori K, Suzuki T, Miyazawa K, Nishiyama S, Yamamura S. Tetrahedron Lett. 1993;34:4981–4984. b) 1:1.8 E:Z in ref. 37b; c) 1:1.6 and 1:2.0 E:Z in ref. 37c; d) selected unpublished results from the Wender group can be found in the Supporting Information (Figure S8).
- 41.HWE selectivity has been achieved through use of chiral phosphonoacetates. This strategy was used in the Evans, Yamamura, Keck, Wender, and Krische total synthetic efforts (see ref. 37), though no fluorinated phosphonoacetates have been used with such a method.
- 42.Notably, a model system derived from hydrocinnamaldehyde and isobutyraldehyde afforded the expected 1:1 dr upon olefination with phosphonoacetate 20.
- 43.Wender P, Billingsley K. Synthesis. 2013;45:1815–1824. doi: 10.1055/s-0033-1338860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Available in four steps from acrolein and benzyl alcohol, see: Wender P, Baryza J, Brenner S, DeChristopher B, Loy B, Schrier A, Verma V. Proc. Natl. Acad. Sci. 2011;108:6721–6726. doi: 10.1073/pnas.1015270108.
- 45.Jasti R, Anderson C, Rycknovsky S. J. Am. Chem. Soc. 2005;127:9939–9945. doi: 10.1021/ja0518326. [DOI] [PubMed] [Google Scholar]
- 46.Binding assay protocol taken from reference 10.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.