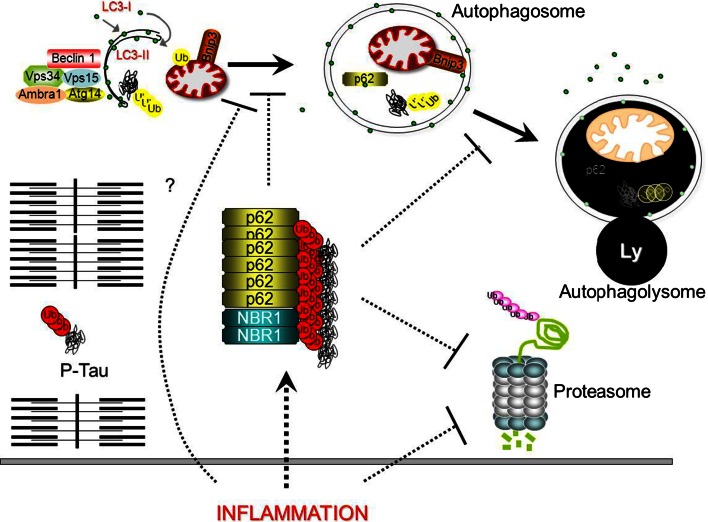
Fig. 3.
In sIBM, misfolded proteins start to accumulate into aggregates that are positive for P62 and NBR1. The presence of P62-positive aggregates is a consequence of impaired protein clearance of proteasome and autophagy/lysosome (Ly) systems that further impacts on their activities. Overload of the autophagy system with subsequent exhaustion, aspecific absorption and sequestration of the proteasome on the surface of inclusions describe how aggregates negatively affect these systems. Autophagy is characterized by membranes that are committed to growth, thus becoming double membrane vesicles, named autophagosome, that surround a ‘portion’ of cytoplasm, organelles, glycogen and protein aggregates. Autophagy is triggered by the activation of a regulatory complex (containing Vps34, Beclin 1, Vps15, Ambra1, Atg14) that induces LC3 recruitment to the nascent autophagosome (isolation membrane). Selective removal of organelles including mitochondria (mitophagy, a specific form of autophagy) requires several signals. For example in mitophagy, Bnip3 factors are recruited on damaged mitochondria and by binding LC3 allow the recruitment of the vesicle on the surface of the altered mitochondria. Proteins that are committed for lysosomal degradation are labelled by polyubiquitin chains and delivered to the autophagosome by the p62/NBR1 scaffold proteins that bind LC3. Finally, upon autophagosome fusion with lysosome the cargo is destroyed and constituents are recycled by the cell to rebuild organelles/proteins or for energy purposes. Whether inflammation triggers protein misfolding aggregation or blocks the autophagy and proteasome system is still an open issue and will certainly be subject of future studies. Dotted lines represent unknown mechanisms (Ub = ubiquitin)