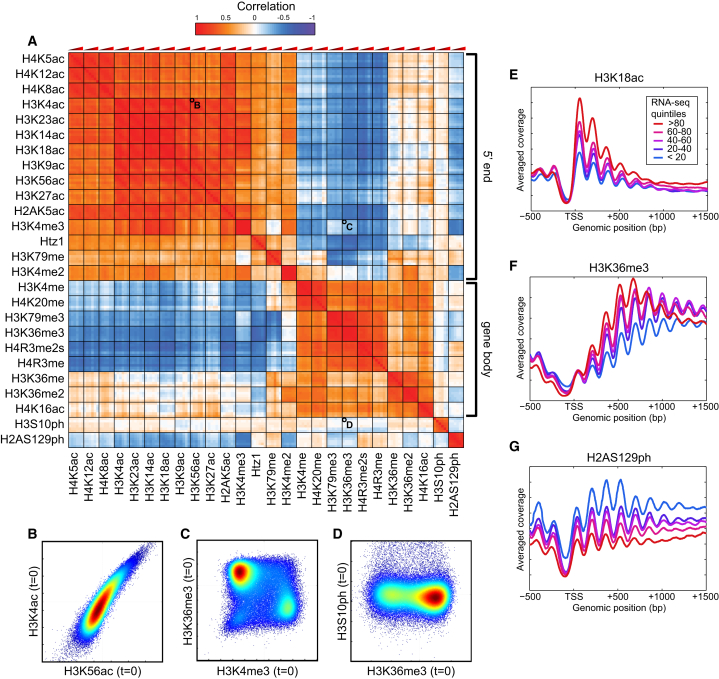
Figure 2.
Characterization of Histone Modification Patterns during Mid-Log Growth
(A) Correlation matrix for 26 histone modifications. For each modification, six time points are arranged from t = 0 to t = 60 from left to right.
(B–D) Scatterplots for strongly correlated (B), uncorrelated (C), and anticorrelated (D) pairs of modifications. Each scatterplot compares levels of the two modifications, normalized to nucleosome occupancy, for 66,360 individual nucleosomes in the yeast genome at t = 0. Colors indicate density.
(E–G) Metagene profiles for exemplary histone modifications. For each modification, data were aligned by the transcription start site (TSS) of annotated genes, grouped according to transcription rate (Churchman and Weissman, 2011).