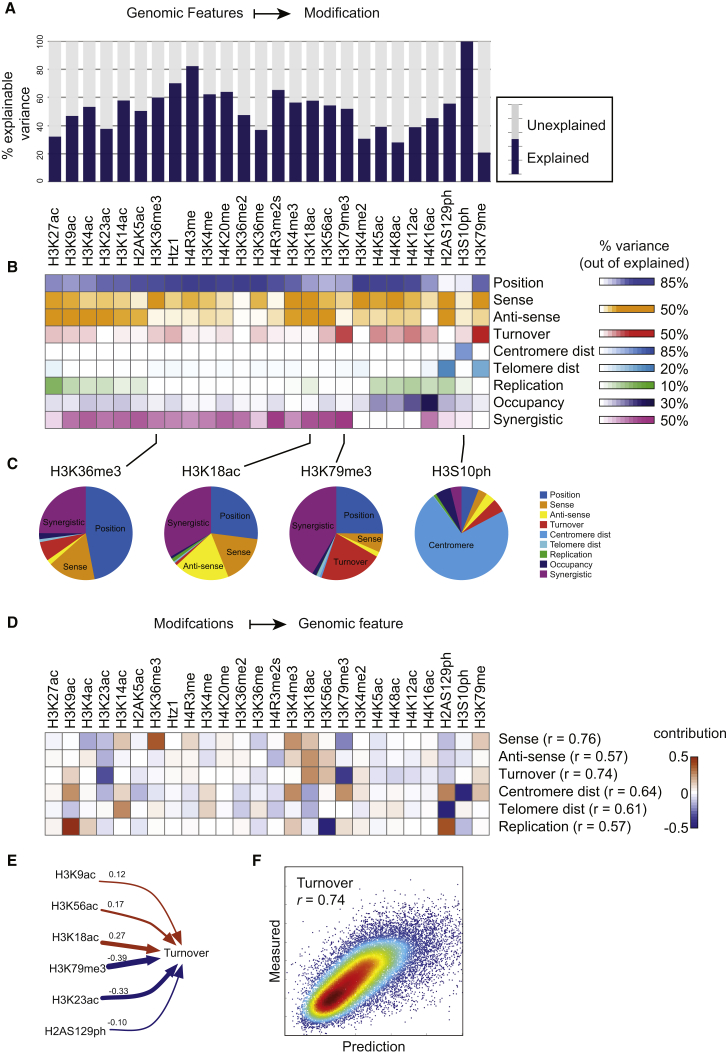
Figure 3.
Determinants of the Steady-State Modification Landscape
(A) Predicting modification data from genomic features. A model incorporating genomic features (sense and antisense transcription (Churchman and Weissman, 2011), nucleosome turnover rate (Dion et al., 2007), distance from centromere and telomere, replication timing (Raghuraman et al., 2001), and nucleosome position) predicts genomic patterns of all 26 histone marks. Plot shows the percent of signal explained per histone modification (see Figure S2B).
(B) Contribution of genomic processes to explanatory power of the model. Heatmap shows the percentage of explained signal that is lost when a given process is removed from the model. Synergistic refers to remaining explained variance not lost upon removing any single feature.
(C) Pie charts showing the variance explained by different aspects of the model for the indicated modifications.
(D) Predicting genomic features from modification data. For each entry, the heatmap shows the sparse linear regression coefficient for the mark in question.
(E) Turnover model parameters from (D) are shown here in numeric form.
(F) Turnover model accurately captures turnover rates genome-wide. Model predictions (x axis) are scatterplotted against experimental turnover data (y axis).