Figure 4.
Dynamics of Histone Modifications during the Stress Response
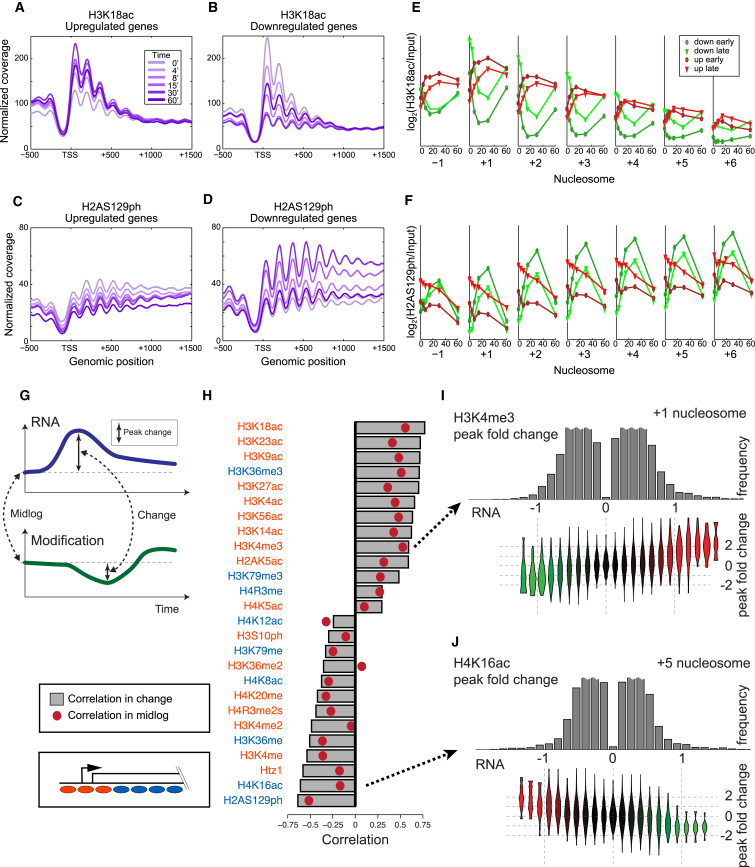
(A and B) Metagenes showing levels of the transcription-correlated H3K18ac mark, averaged for upregulated (A) or downregulated (B) genes in response to diamide stress.
(C and D) As in (A) and (B), for the repression-correlated H2AS129ph modification.
(E and F) Dynamics of H3K18ac (E) or H2AS129ph (F) changes over time are shown averaged for various nucleosome positions along a gene body—the −1, +1, +2, etc. nucleosomes—as indicated. For each nucleosome, time course data for the modification in question are averaged for genes upregulated, or downregulated, relatively rapidly or slowly (Experimental Procedures).
(G) Schematic of approach to correlations between histone modification dynamics and transcriptional dynamics.
(H) Correlations calculated as shown in (G), with red dots showing mid-log correlations, and gray bars showing correlations between diamide-induced change in modification and change in transcription.
(I and J) The correspondence between modification changes during diamide stress and transcription changes. In each case, a specific nucleosome location (+1, +5) is indicated. (Top panel) Histogram of the maximal change in the listed modification in response to diamide. (Bottom panel) Violin plots of changes in mRNA abundance for the genes carrying the nucleosomes in the bins above.