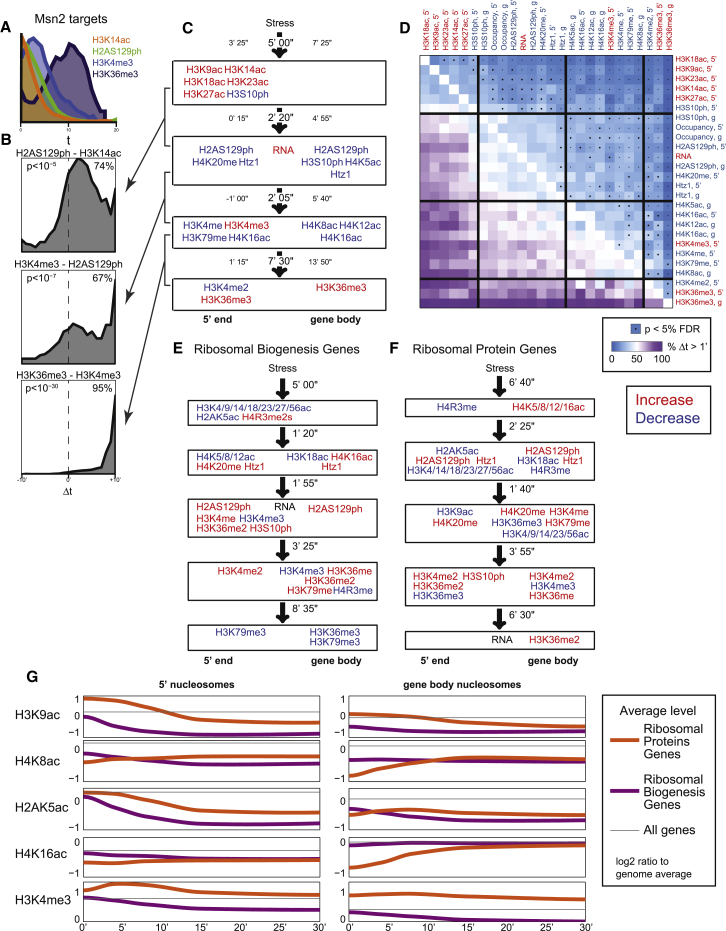
Figure 7.
Cascades of Chromatin Events Differ between Gene Sets
(A) Distribution of t1/2 values for the four indicated marks for all MSN2-induced genes.
(B) Gene-by-gene analysis for differences in modification onset times. The distribution of the difference in t1/2 is calculated for all individual genes in the MSN2-dependent gene set for the indicated modification pair.
(C) Four “epochs” in the MSN2 induction cascade. Groups of histone modification changes: modifications in each group roughly co-occur, but differ significantly in timing in pairwise comparisons from the other groups. For each box, the mean and 25th and 75th percentile values are shown for the distribution of differences in t1/2 between modifications in adjacent boxes.
(D) Heatmap showing all pairwise comparisons for MSN2-dependent upregulated genes. Each row/column represents a modification and a genic location (5′ end, or gene body) that changes coherently for MSN2-upregulated genes. Heavy lines show demarcation for the boxes summarized in (C).
(E and F) Summary diagrams, as in (C), for RiBi genes and RPGs, as indicated.
(G) Interpolated time course data for RiBi genes and RPGs for 30 min of stress response. The shown modification levels are averages, for genes in each group, of the log2 ratio to genome-wide mean at t = 0.