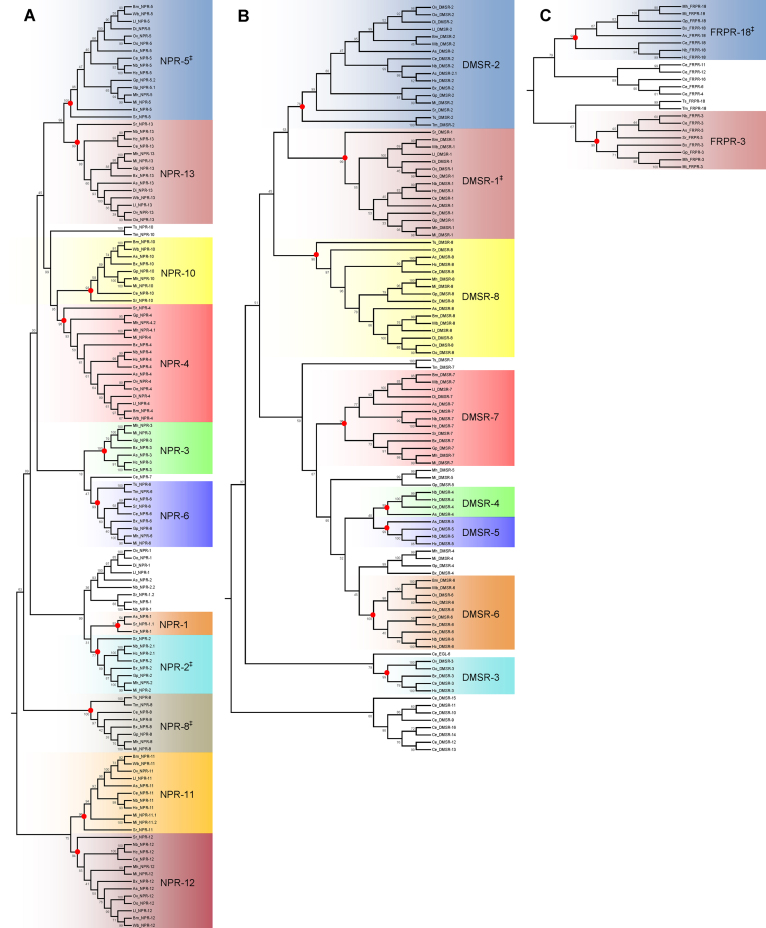
Fig. 4.
Maximum-likelihood phylogenies of putative FLP-GPCRs from parasitic nematodes.
The phylogenies reveal 13 putative FLP-GPCRs that are highly conserved in nematode parasites (see Fig. 3). Entries are identified as per previous designation [28] and prefixes which indicate species: Ts, denotes Trichinella spiralis; Tm, denotes Trichuris muris; Bm, denotes Brugia malayi; Wb, denotes Wuchereria bancrofti; Di, denotes Dirofilaria immitis; Ov, denotes Onchocerca volvulus; Oo, denotes Onchocerca ochengi; Ll, denotes Loa loa; As, denotes Ascaris suum; Ac, denotes Ancylostoma caninum; Nb, denotes Nippostrongylus brasiliensis; Hc, denotes Haemonchus contortus; Ce, denotes Caenorhabditis elegans; Sr, denotes Strongyloides ratti; Bx, denotes Bursaphelenchus xylophilus; Gp, denotes Globodera pallida; Mh, denotes Meloidogyne hapla; Mi, denotes Meloidogyne incognita. Each phylogeny (A-C) represents a single C. elegans FLP-GPCR cluster identified in Fig. 3: (A) denotes the phylogeny of parasitic nematode FLP-GPCR homologues of the C. elegans predicted proteins present in Fig. 3 cluster 1; (B) denotes the phylogeny of parasitic nematode FLP-GPCR homologues of the C. elegans predicted proteins present in Fig. 3 cluster 2; (C) denotes the phylogeny of parasitic nematode FLP-GPCR homologues of the C. elegans predicted proteins present in Fig. 3 cluster 3. FLP-GPCR cluster nodes supported by a bootstrap analysis value of >70% are identified by red dots. Clusters are shaded and named according to the relevant C. elegans orthologue. Bootstrap values are shown as percentages. ‡Note that NPR-2, NPR-5, NPR-8, DMSR-1 and FRPR-18 isoforms were not delineated in the parasite species. In addition only the longest C. elegans FLP-/putative FLP-receptor isoforms were included in the phylogenetic analyses such that Ce_NPR-2 represents isoform a; Ce_NPR-5 represents isoform b; Ce_NPR-8 represents isoform b, Ce_DMSR-1 represents isoform a; Ce_EGL-6 represents isoform a; Ce_FRPR-18 represents isoform b.