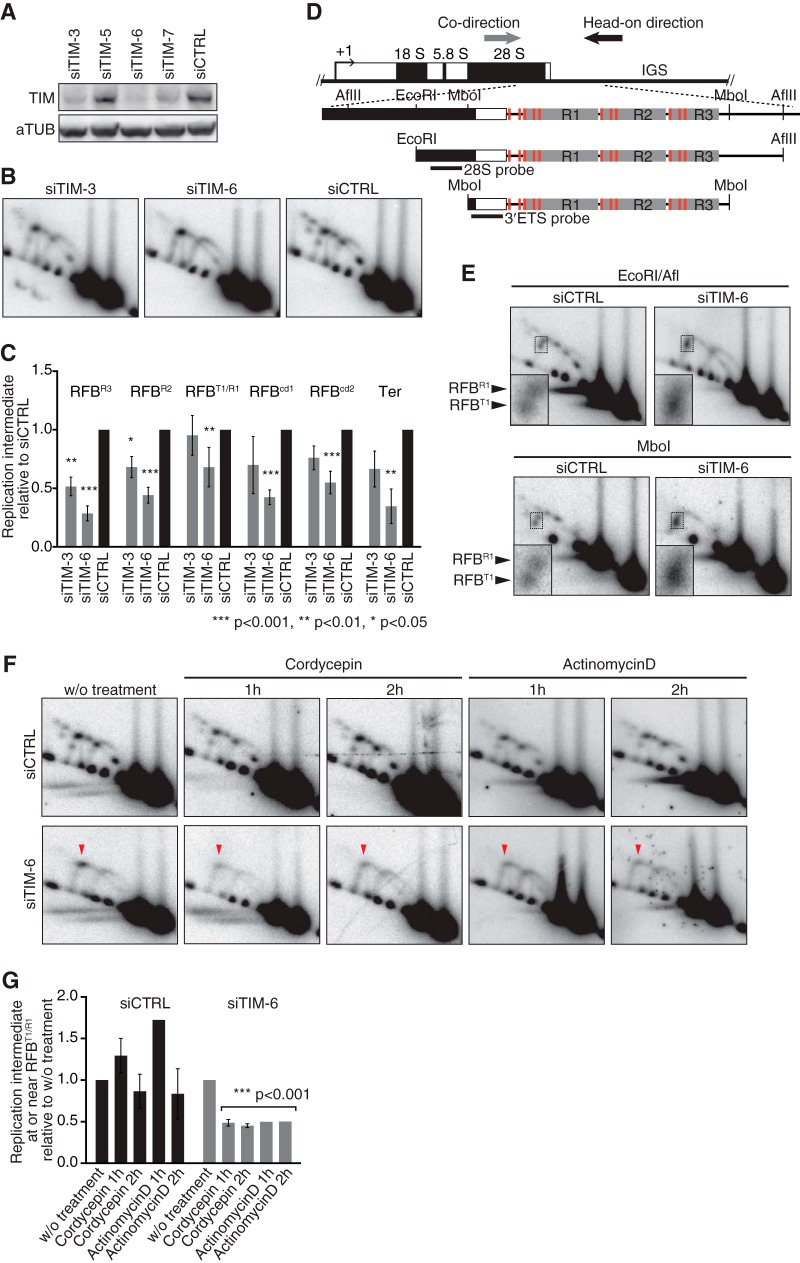
FIG 6.
TIM is required for replication fork arrest at RFBs to coordinate the progression of replication with transcription activity. (A) Cellular depletion of TIM in HeLa cells was verified by Western blotting. (B) 2-D gel analysis of AflII fragments from TIM-depleted cells 96 h after transfection. (C) Quantification of replication intermediates detected as shown in panel B was as described in the legend to Fig. 3. The mean results and SD from independent experiments (n = 3 for siTIM-3 and n = 7 for siTIM-6 and siCTRL) are presented. (D) Diagram of class I rDNA fragments with restriction sites for AflII, MboI, and EcoRI and the positions of probes indicated. (E) 2-D gel analysis of EcoRI-AflII and MboI fragments, which were hybridized to the 28S probe and the 3′ETS probe, respectively. Insets show magnifications of the Y forks accumulating around RFBR1 and RFBT1. (F) Effect of transcription inhibition on replication. 2-D gel analyses of AflII fragments from cells that were treated with cordycepin (50 μM) or actinomycin D (50 ng/ml) for the indicated periods. Replication intermediates were visualized with the 28S probe. Arrowheads indicate transcription-dependent Y fork accumulation. (G) Quantification of replication intermediates at or near RFBR1/T1 detected as shown in panel F. Values relative to that of the signal in cells without (w/o) treatment are shown. For siTIM-6-treated cells, all values from inhibitor-treated cells were combined to calculate the P values using a one-sample t test with hypothetical mean of 1.