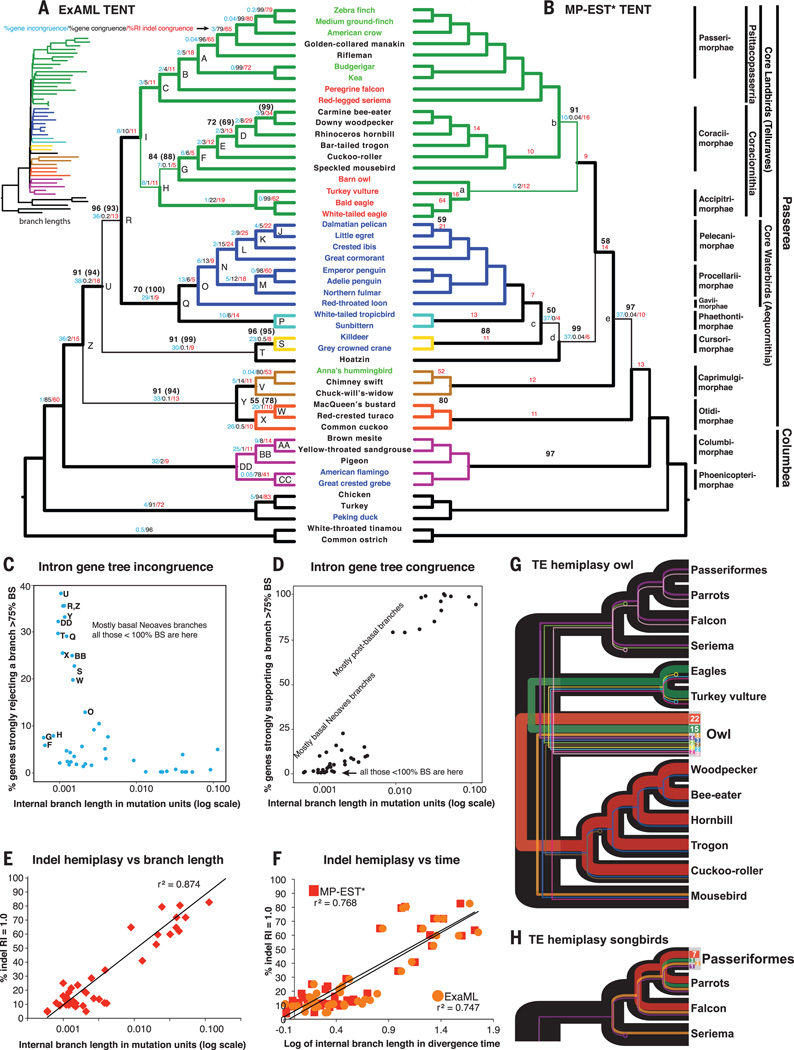
Fig. 3. Evidence of ILS.
(A) Cladogram of ExaML TENT avian species tree, annotated for nodes from Fig. 2 (letters), for branches with less than 100% BS without and with (parentheses) third codon positions, for strong (>75% BS) intron gene tree incongruence and congruence, and for indel congruence on all branches (except the root). Thin branch lines represent those not present in the MP-EST* TENT of (B). (Inset) ExaML branch lengths in substitution units (expanded view in fig. S7). Color coding of branches and species is as in Fig. 1. (B) Cladogram of MP-EST* TENT species tree, annotated similarly as in the ExaML TENT in (A). Thin branch lines represent those not present in the ExaML TENT of (A). (C) Percent of intron gene trees rejecting (≥75% BS) branches in the ExaML TENT species tree relative to branch lengths. Letters denote nodes in (A) that either have less than 100% support or are different from the MP-EST* TENT in (B). (D) Percent of intron gene trees supporting (≥75% BS) branches in the ExaML TENT species tree relative to branch lengths. (E) Indel hemiplasy [the inverse of percent of retention index (RI) = 1.0 indels that support the branch; see SM9] correlated with ExaML TENT branch length (log transformed). r2, correlation coefficient. (F) Indel hemiplasy correlated with ExaML and MP-EST TENT internal branch divergence times in millions of years (log transformed). Plotting with internal branch times was necessary to compare both trees (SM9). (G) TE hemiplasy with owls among the core landbirds. Line color, shared TE tree topology; line thickness, relative proportion of TEs that support a specific topology (total numbers shown in the owl node). Circles at end of lines indicate loss of the TE allele in that species after ILS, as the sequence assembly contains an empty TE insertion site (SM10). Only topologies with two or more TEs are shown. (H) TE hemiplasy with songbirds among the core landbirds.