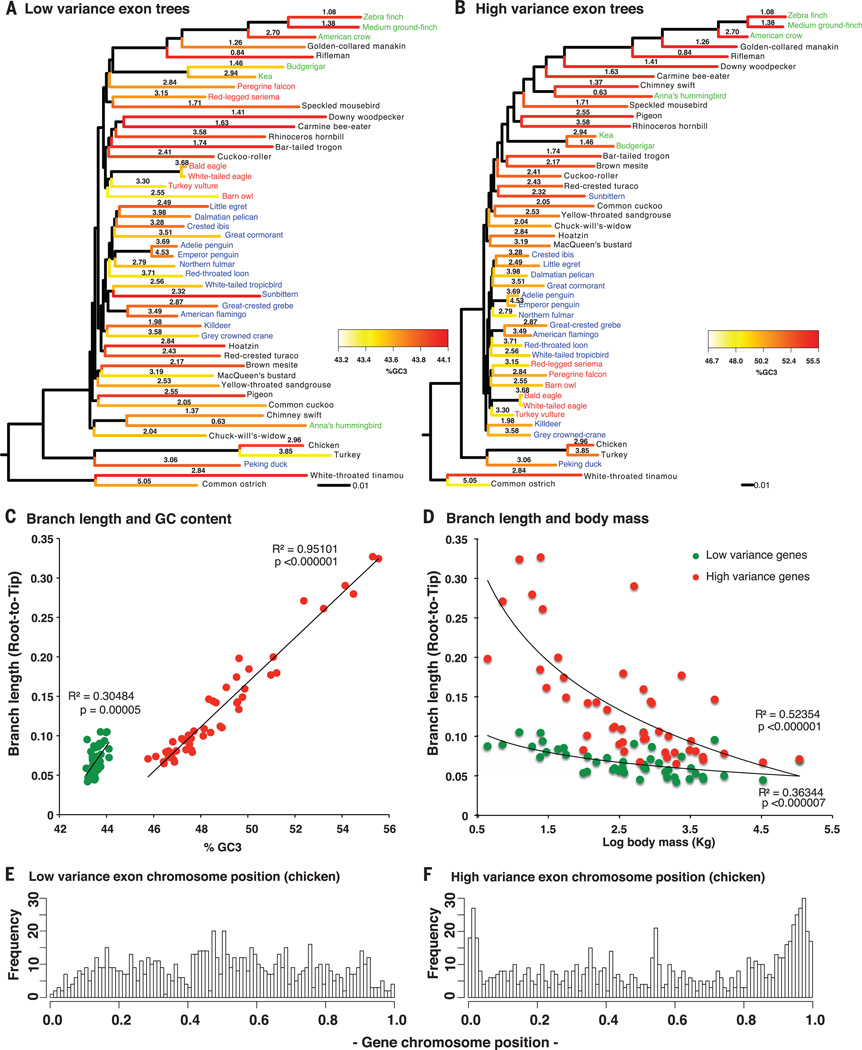
Fig. 6. Life history incongruence in protein-coding trees.
(A) Species tree inferred from low-base composition variance exons (n = 830 genes) graphed with branch length, third codon position GC (GC3) content (heatmap), and log of body mass (numbers on branches). (B) Species tree inferred from high-base composition variance exons (n = 830 genes), graphed similarly as in (A). The %GC3 scale is higher and ~10 times wider for the high-variance genes, and the branch lengths are ~3 times longer [black scales at the bottom of (A) and (B)]. Color coding of species’ names is as in Fig. 1. Cladograms of trees in (A) and (B) are in figs. S16, A and B. (C and D) Correlations of branch length with GC content (C) and body mass (D) of the low-variance and high-variance exons. Correlations were still significant after independent contrast analyses for phylogenetic relationships (SM11). (E and F) Relative chromosome positions of the low-variance (E) and high-variance (F) exons normalized between 0 and 1 for all chicken chromosomes and separated into 100 bins (bars). The height of each bar represents the number of genes in that specific relative location. The two distributions in (E) and (F) are significantly different (P < 2.2 × 10−16, Wilcoxon rank sum test on grouped values). For further details, see SM11.