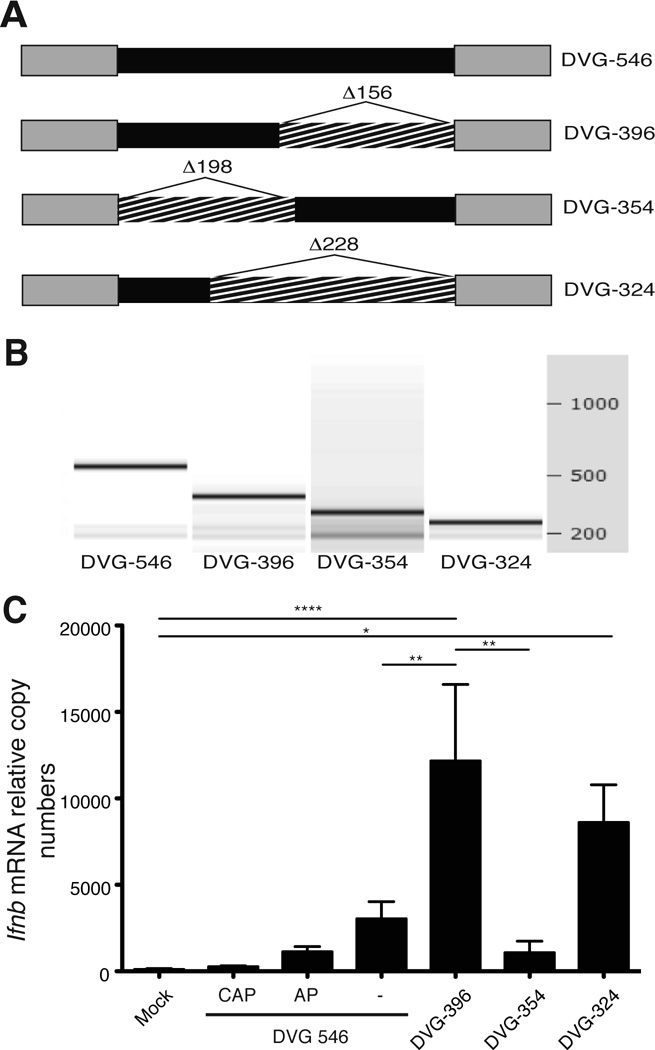
Figure 4. Naked DVG-derived ivtRNA preserves immunostimulatory activity.
(A) Representation of deletion mutants of the DVG genome. Deletions (hatched) were performed in the internal sequence (black) without compromising the DVG complementary ends (grey). (B) The electrophoretic analysis for each ivtRNA was performed in an Agilent's 2100 Bioanalyzer. (C) LLC-MK2 cells were transfected with DVG-546, capped DVG-546 (CAP), DVG-546 treated with alkaline phosphatase (AP) or the different mutants and 6 h post-transfection cells were harvested and total cellular RNA was extracted to determine expression of Ifnb mRNA by RT-qPCR. The experiment was independently repeated three times. Each assay was performed in triplicates. Data corresponds to the average of all experiments (total n = 3–8/group). p=0.0001 by one-way ANOVA with Bonferroni post-hoc test. Significance after Bonferroni post-hoc test among different conditions is indicated in the graphs as ****=p<0.0001, **=p<0.01, and **=p<0.05. Bonferroni denominator = 3. Data are expressed as copy numbers relative to the housekeeping genes Actb and Tuba1b.